Depression of oncogenecity by dephosphorylating and degrading BCR-ABL
- PMID: 27926512
- PMCID: PMC5356883
- DOI: 10.18632/oncotarget.13754
Depression of oncogenecity by dephosphorylating and degrading BCR-ABL
Abstract
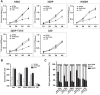
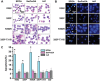
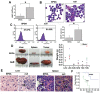
Aberrant phosphorylation and overexpression of BCR-ABL fusion protein are responsible for the main pathogenesis in chronic myeloid leukemia (CML). Phosphorylated BCR-ABL Y177 recruits GRB2 adaptor and triggers leukemic RAS-MAPK and PI3K-AKT signals. In this study, we engineered a SPOA system to dephosphorylate and degrade BCR-ABL by targeting BCR-ABL Y177. We tested its effect on BCR-ABL phosphorylation and expression, as well as cell proliferation and apoptosis in CML cells. We found that SPOA remarkably dephosphorylated BCR-ABL Y177, prevented GRB2 recruitment, and uncoupled RAS-MAPK and PI3K-AKT signals. Meanwhile, SPOA degraded BCR-ABL oncoprotein in ubiquitin-independent manner and depressed the signal transduction of STAT5 and CRKL by BCR-ABL. Furthermore, SPOA inhibited proliferation and induced apoptosis in CML cells and depressed the oncogenecity of K562 cells in mice. These results provide evidence that dephosphorylating and degrading oncogenic BCR-ABL offer an alternative CML therapy.
Keywords: BCR-ABL; Y177; chronic myeloid leukemia; ornithine decarboxylase; protein tyrosine phosphatase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Growth of chronic myeloid leukemia cells is inhibited by infection with Ad-SH2-HA adenovirus that disrupts Grb2-Bcr-Abl complexes.Oncol Rep. 2011 May;25(5):1381-8. doi: 10.3892/or.2011.1197. Epub 2011 Mar 1. Oncol Rep. 2011. PMID: 21369701
-
Induction of apoptosis in imatinib sensitive and resistant chronic myeloid leukemia cells by efficient disruption of bcr-abl oncogene with zinc finger nucleases.J Exp Clin Cancer Res. 2018 Mar 20;37(1):62. doi: 10.1186/s13046-018-0732-4. J Exp Clin Cancer Res. 2018. PMID: 29554925 Free PMC article.
-
Blockade of Y177 and Nuclear Translocation of Bcr-Abl Inhibits Proliferation and Promotes Apoptosis in Chronic Myeloid Leukemia Cells.Int J Mol Sci. 2017 Mar 2;18(3):537. doi: 10.3390/ijms18030537. Int J Mol Sci. 2017. PMID: 28257089 Free PMC article.
-
Chronic Myeloid Leukemia in the Era of Tyrosine Kinase Inhibitors: An Evolving Paradigm of Molecularly Targeted Therapy.Mol Diagn Ther. 2016 Aug;20(4):315-33. doi: 10.1007/s40291-016-0208-1. Mol Diagn Ther. 2016. PMID: 27220498 Review.
-
Growth factor independence and BCR/ABL transformation: promise and pitfalls of murine model systems and assays.Leukemia. 1999 Aug;13(8):1200-6. doi: 10.1038/sj.leu.2401467. Leukemia. 1999. PMID: 10450747 Review.
Cited by
-
RanBP3 Regulates Proliferation, Apoptosis and Chemosensitivity of Chronic Myeloid Leukemia Cells via Mediating SMAD2/3 and ERK1/2 Nuclear Transport.Front Oncol. 2021 Aug 24;11:698410. doi: 10.3389/fonc.2021.698410. eCollection 2021. Front Oncol. 2021. PMID: 34504783 Free PMC article.
-
Mathematical models of amino acid panel for assisting diagnosis of children acute leukemia.J Transl Med. 2019 Jan 23;17(1):38. doi: 10.1186/s12967-019-1783-9. J Transl Med. 2019. PMID: 30674317 Free PMC article.
-
Regulative Loop between β-catenin and Protein Tyrosine Receptor Type γ in Chronic Myeloid Leukemia.Int J Mol Sci. 2020 Mar 26;21(7):2298. doi: 10.3390/ijms21072298. Int J Mol Sci. 2020. PMID: 32225105 Free PMC article.
-
Current Views on the Interplay between Tyrosine Kinases and Phosphatases in Chronic Myeloid Leukemia.Cancers (Basel). 2021 May 12;13(10):2311. doi: 10.3390/cancers13102311. Cancers (Basel). 2021. PMID: 34065882 Free PMC article. Review.
References
-
- Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 1986;47:277–284. - PubMed
-
- Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. - PubMed
-
- ten Hoeve J, Kaartinen V, Fioretos T, Haataja L, Voncken JW, Heisterkamp N, Groffen J. Cellular interactions of CRKL, and SH2-SH3 adaptor protein. Cancer Res. 1994;54:2563–2567. - PubMed
-
- Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

