Modeling microelectrode biosensors: free-flow calibration can substantially underestimate tissue concentrations
- PMID: 27927788
- PMCID: PMC5338626
- DOI: 10.1152/jn.00788.2016
Modeling microelectrode biosensors: free-flow calibration can substantially underestimate tissue concentrations
Abstract
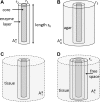
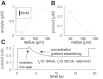
Microelectrode amperometric biosensors are widely used to measure concentrations of analytes in solution and tissue including acetylcholine, adenosine, glucose, and glutamate. A great deal of experimental and modeling effort has been directed at quantifying the response of the biosensors themselves; however, the influence that the macroscopic tissue environment has on biosensor response has not been subjected to the same level of scrutiny. Here we identify an important issue in the way microelectrode biosensors are calibrated that is likely to have led to underestimations of analyte tissue concentrations. Concentration in tissue is typically determined by comparing the biosensor signal to that measured in free-flow calibration conditions. In a free-flow environment the concentration of the analyte at the outer surface of the biosensor can be considered constant. However, in tissue the analyte reaches the biosensor surface by diffusion through the extracellular space. Because the enzymes in the biosensor break down the analyte, a density gradient is set up resulting in a significantly lower concentration of analyte near the biosensor surface. This effect is compounded by the diminished volume fraction (porosity) and reduction in the diffusion coefficient due to obstructions (tortuosity) in tissue. We demonstrate this effect through modeling and experimentally verify our predictions in diffusive environments.NEW & NOTEWORTHY Microelectrode biosensors are typically calibrated in a free-flow environment where the concentrations at the biosensor surface are constant. However, when in tissue, the analyte reaches the biosensor via diffusion and so analyte breakdown by the biosensor results in a concentration gradient and consequently a lower concentration around the biosensor. This effect means that naive free-flow calibration will underestimate tissue concentration. We develop mathematical models to better quantify the discrepancy between the calibration and tissue environment and experimentally verify our key predictions.
Keywords: biosensors; calibration; modeling.
Copyright © 2017 the American Physiological Society.
Figures




References
-
- Baronas R, Ivanauskas F, Kulys J. Mathematical Modeling of Biosensors: an Introduction for Chemists and Mathematicians. New York: Springer, 2009.
-
- Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14: 724–738, 2011. - PubMed
-
- Berti V, Vanzi E, Polito C, Pupi A. Back to the future: the absolute quantification of cerebral metabolic rate of glucose. Clin Trans Imaging 1: 289–296, 2013.
-
- Betz AL. Identification of hypoxanthine transport and xanthine oxidase activity in brain capillaries. J Neurochem 44: 574–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

