Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila
- PMID: 27929060
- PMCID: PMC5155165
- DOI: 10.1038/ncomms13739
Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila
Abstract
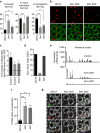
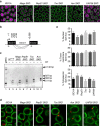
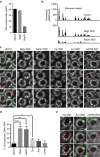
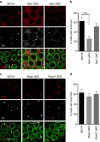
PIWI-interacting RNAs (piRNAs) are effectors of transposable element (TE) silencing in the reproductive apparatus. In Drosophila ovarian somatic cells, piRNAs arise from longer single-stranded RNA precursors that are processed in the cytoplasm presumably within the Yb-bodies. piRNA precursors encoded by the flamenco (flam) piRNA cluster accumulate in a single focus away from their sites of transcription. In this study, we identify the exportin complex containing Nxf1 and Nxt1 as required for flam precursor nuclear export. Together with components of the exon junction complex (EJC), it is necessary for the efficient transfer of flam precursors away from their site of transcription. Indeed, depletion of these components greatly affects flam intra-nuclear transit. Moreover, we show that Yb-body assembly is dependent on the nucleo-cytoplasmic export of flam transcripts. These results suggest that somatic piRNA precursors are thus required for the assembly of the cytoplasmic transposon silencing machinery.
Figures

References
-
- Brennecke J. et al.. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

