Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer
- PMID: 27929064
- PMCID: PMC5155160
- DOI: 10.1038/ncomms13665
Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer
Abstract
Blockade of the epidermal growth factor receptor (EGFR) with the monoclonal antibodies cetuximab or panitumumab is effective in a subset of colorectal cancers (CRCs), but the emergence of resistance limits the efficacy of these therapeutic agents. At relapse, the majority of patients develop RAS mutations, while a subset acquires EGFR extracellular domain (ECD) mutations. Here we find that patients who experience greater and longer responses to EGFR blockade preferentially develop EGFR ECD mutations, while RAS mutations emerge more frequently in patients with smaller tumour shrinkage and shorter progression-free survival. In circulating cell-free tumour DNA of patients treated with anti-EGFR antibodies, RAS mutations emerge earlier than EGFR ECD variants. Subclonal RAS but not EGFR ECD mutations are present in CRC samples obtained before exposure to EGFR blockade. These data indicate that clonal evolution of drug-resistant cells is associated with the clinical outcome of CRC patients treated with anti-EGFR antibodies.
Conflict of interest statement
A.S.-B. is a member of advisory boards for Amgen, Bayer, Lilly and Sanofi. S.S. is a member of advisory boards for Amgen, Roche, Bayer, Sanofi, Eli Lilly and Merck. The remaining authors declare no competing financial interests.
Figures

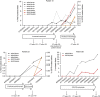
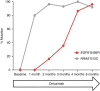
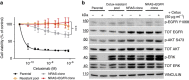

References
-
- Burrell R. A., McGranahan N., Bartek J. & Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

