Identification of a CD133-CD55- population functions as a fetal common skeletal progenitor
- PMID: 27929130
- PMCID: PMC5144148
- DOI: 10.1038/srep38632
Identification of a CD133-CD55- population functions as a fetal common skeletal progenitor
Abstract
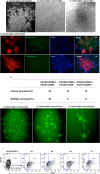
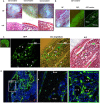
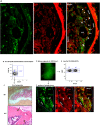
In this study, we identified a CD105+CD90.1-CD133-CD55- (CD133-CD55-) population in the fetal skeletal element that can generate bone and bone marrow. Besides osteoblasts and chondrocytes, the CD133-CD55- common progenitors can give rise to marrow reticular stromal cells and perivascular mesenchymal progenitors suggesting they function as the fetal common skeletal progenitor. Suppression of CXCL12 and Kitl expression in CD133-CD55- common progenitors severely disrupted the BM niche formation but not bone generation. Thus, CD133-CD55- common progenitors are the main source of CXCL12 and Kitl producing cells in the developing marrow.
Figures




Similar articles
-
C-KIT Expression Distinguishes Fetal from Postnatal Skeletal Progenitors.Stem Cell Reports. 2020 Apr 14;14(4):614-630. doi: 10.1016/j.stemcr.2020.03.001. Epub 2020 Mar 26. Stem Cell Reports. 2020. PMID: 32220331 Free PMC article.
-
Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation.Nature. 2014 Apr 24;508(7497):536-40. doi: 10.1038/nature13071. Epub 2014 Mar 2. Nature. 2014. PMID: 24590069
-
CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance.Nature. 2013 Mar 14;495(7440):227-30. doi: 10.1038/nature11926. Epub 2013 Feb 24. Nature. 2013. PMID: 23434756 Free PMC article.
-
[Bone and Stem Cells. Bone marrow microenvironment niches for hematopoietic stem and progenitor cells].Clin Calcium. 2014 Apr;24(4):517-26. Clin Calcium. 2014. PMID: 24681497 Review. Japanese.
-
Hematopoietic Multipotent Progenitors and Plasma Cells: Neighbors or Roommates in the Mouse Bone Marrow Ecosystem?Front Immunol. 2021 Apr 15;12:658535. doi: 10.3389/fimmu.2021.658535. eCollection 2021. Front Immunol. 2021. PMID: 33936091 Free PMC article. Review.
Cited by
-
Single-cell RNA landscape of the osteoimmunology microenvironment in periodontitis.Theranostics. 2022 Jan 1;12(3):1074-1096. doi: 10.7150/thno.65694. eCollection 2022. Theranostics. 2022. PMID: 35154475 Free PMC article.
-
Different phenotypes and chondrogenic responses of human menstrual blood and bone marrow mesenchymal stem cells to activin A and TGF-β3.Stem Cell Res Ther. 2021 Apr 29;12(1):251. doi: 10.1186/s13287-021-02286-w. Stem Cell Res Ther. 2021. PMID: 33926568 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

