miR-17∼92 family clusters control iNKT cell ontogenesis via modulation of TGF-β signaling
- PMID: 27930306
- PMCID: PMC5187732
- DOI: 10.1073/pnas.1612024114
miR-17∼92 family clusters control iNKT cell ontogenesis via modulation of TGF-β signaling
Abstract
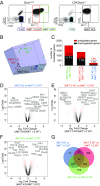
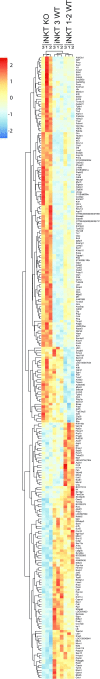
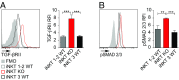
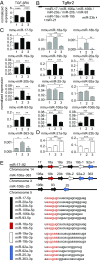
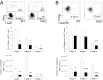
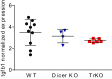
Invariant natural killer T cells (iNKT) cells are T lymphocytes displaying innate effector functions, acquired through a distinct thymic developmental program regulated by microRNAs (miRNAs). Deleting miRNAs by Dicer ablation (Dicer KO) in thymocytes selectively impairs iNKT cell survival and functional differentiation. To unravel this miRNA-dependent program, we systemically identified transcripts that were differentially expressed between WT and Dicer KO iNKT cells at different differentiation stages and predicted to be targeted by the iNKT cell-specific miRNAs. TGF-β receptor II (TGF-βRII), critically implicated in iNKT cell differentiation, was found up-regulated in iNKT Dicer KO cells together with enhanced TGF-β signaling. miRNA members of the miR-17∼92 family clusters were predicted to target Tgfbr2 mRNA upon iNKT cell development. iNKT cells lacking all three miR-17∼92 family clusters (miR-17∼92, miR-106a∼363, miR-106b∼25) phenocopied both increased TGF-βRII expression and signaling, and defective effector differentiation, displayed by iNKT Dicer KO cells. Consistently, genetic ablation of TGF-β signaling in the absence of miRNAs rescued iNKT cell differentiation. These results elucidate the global impact of miRNAs on the iNKT cell developmental program and uncover the targeting of a lineage-specific cytokine signaling by miRNAs as a mechanism regulating innate-like T-cell development and effector differentiation.
Keywords: CD1d; NKT cells; TGF-β; development; miRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Dicer-dependent microRNA pathway controls invariant NKT cell development.J Immunol. 2009 Aug 15;183(4):2506-12. doi: 10.4049/jimmunol.0901361. Epub 2009 Jul 22. J Immunol. 2009. PMID: 19625646
-
Invariant NKT cell development and function in microRNA-223 knockout mice.Int Immunopharmacol. 2011 May;11(5):561-8. doi: 10.1016/j.intimp.2010.11.004. Epub 2010 Nov 19. Int Immunopharmacol. 2011. PMID: 21094288
-
Loss of microRNAs in thymus perturbs invariant NKT cell development and function.Cell Mol Immunol. 2010 Nov;7(6):447-53. doi: 10.1038/cmi.2010.49. Epub 2010 Sep 20. Cell Mol Immunol. 2010. PMID: 20852654 Free PMC article.
-
T Cell Receptor Expression Timing and Signal Strength in the Functional Differentiation of Invariant Natural Killer T Cells.Front Immunol. 2019 Apr 26;10:841. doi: 10.3389/fimmu.2019.00841. eCollection 2019. Front Immunol. 2019. PMID: 31080448 Free PMC article. Review.
-
microRNA dynamic expression regulates invariant NKT cells.Cell Mol Life Sci. 2021 Aug;78(16):6003-6015. doi: 10.1007/s00018-021-03895-7. Epub 2021 Jul 8. Cell Mol Life Sci. 2021. PMID: 34236444 Free PMC article. Review.
Cited by
-
A feed-forward regulatory network lncPCAT1/miR-106a-5p/E2F5 regulates the osteogenic differentiation of periodontal ligament stem cells.J Cell Physiol. 2019 Nov;234(11):19523-19538. doi: 10.1002/jcp.28550. Epub 2019 Apr 17. J Cell Physiol. 2019. Retraction in: J Cell Physiol. 2023 Feb;238(2):476. doi: 10.1002/jcp.30960. PMID: 30997692 Free PMC article. Retracted.
-
Prognostic value of miR17-92 family in patients with digestive system cancers: a systematic review and meta-analysis.Cancer Manag Res. 2019 Jan 22;11:1043-1058. doi: 10.2147/CMAR.S189113. eCollection 2019. Cancer Manag Res. 2019. PMID: 30774432 Free PMC article. Review.
-
Prognostic role of miR-17-92 family in human cancers: evaluation of multiple prognostic outcomes.Oncotarget. 2017 Jul 8;8(40):69125-69138. doi: 10.18632/oncotarget.19096. eCollection 2017 Sep 15. Oncotarget. 2017. PMID: 28978185 Free PMC article. Review.
-
MicroRNAs in cartilage development and dysplasia.Bone. 2020 Nov;140:115564. doi: 10.1016/j.bone.2020.115564. Epub 2020 Jul 31. Bone. 2020. PMID: 32745689 Free PMC article. Review.
-
The Emerging Role of Epigenetics in Autoimmune Thyroid Diseases.Front Immunol. 2017 Apr 7;8:396. doi: 10.3389/fimmu.2017.00396. eCollection 2017. Front Immunol. 2017. PMID: 28439272 Free PMC article. Review.
References
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
-
- Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–117. - PubMed
-
- Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

