Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis
- PMID: 27930336
- PMCID: PMC5187681
- DOI: 10.1073/pnas.1618258113
Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis
Abstract
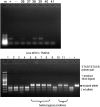
Despite the major role of chitin biosynthesis inhibitors such as benzoylureas (BPUs) in the control of pests in agricultural and public health for almost four decades, their molecular mode of action (MoA) has in most cases remained elusive. BPUs interfere with chitin biosynthesis and were thought to interact with sulfonylurea receptors that mediate chitin vesicle transport. Here, we uncover a mutation (I1042M) in the chitin synthase 1 (CHS1) gene of BPU-resistant Plutella xylostella at the same position as the I1017F mutation reported in spider mites that confers etoxazole resistance. Using a genome-editing CRISPR/Cas9 approach coupled with homology-directed repair (HDR) in Drosophila melanogaster, we introduced both substitutions (I1056M/F) in the corresponding fly CHS1 gene (kkv). Homozygous lines bearing either of these mutations were highly resistant to etoxazole and all tested BPUs, as well as buprofezin-an important hemipteran chitin biosynthesis inhibitor. This provides compelling evidence that BPUs, etoxazole, and buprofezin share in fact the same molecular MoA and directly interact with CHS. This finding has immediate effects on resistance management strategies of major agricultural pests but also on mosquito vectors of serious human diseases such as Dengue and Zika, as diflubenzuron, the standard BPU, is one of the few effective larvicides in use. The study elaborates on how genome editing can directly, rapidly, and convincingly elucidate the MoA of bioactive molecules, especially when target sites are complex and hard to reconstitute in vitro.
Keywords: CRISPR/Cas9; benzoylureas; insecticide resistance; mosquito control; resistance management.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

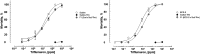






References
-
- World Health Organization 2016 World Malaria Report, 2015 (WHO, Geneva, Switzerland). Available at www.who.int/mediacentre/factsheets/fs387/en/. Accessed October 31, 2016.
-
- Food and Agriculture Organization of the United Nations 2010 How to Feed the World in 2050. Available at www.fao.org/fileadmin/templates/wsfs/docs/expert_paper/How_to_Feed_the_W.... Accessed October 31, 2016.
-
- Talekar NS, Shelton AM. Biology, ecology and management of the diamondback moth. Annu Rev Entomol. 1993;38:275–301. - PubMed
-
- Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pestic Biochem Physiol. 2015;121:122–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

