Transcriptome profiling reveals differential gene expression of detoxification enzymes in a hemimetabolous tobacco pest after feeding on jasmonate-silenced Nicotiana attenuata plants
- PMID: 27931186
- PMCID: PMC5146904
- DOI: 10.1186/s12864-016-3348-0
Transcriptome profiling reveals differential gene expression of detoxification enzymes in a hemimetabolous tobacco pest after feeding on jasmonate-silenced Nicotiana attenuata plants
Abstract
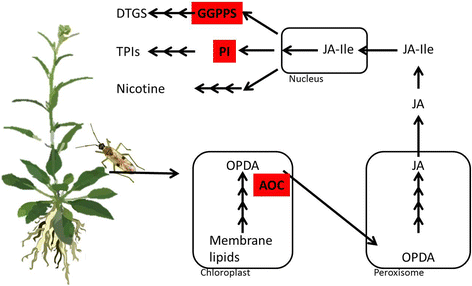
Background: The evolutionary arms race between plants and insects has driven the co-evolution of sophisticated defense mechanisms used by plants to deter herbivores and equally sophisticated strategies that enable phytophagous insects to rapidly detoxify the plant's defense metabolites. In this study, we identify the genetic determinants that enable the mirid, Tupiocoris notatus, to feed on its well-defended host plant, Nicotiana attenuata, an outstanding model for plant-insect interaction studies.
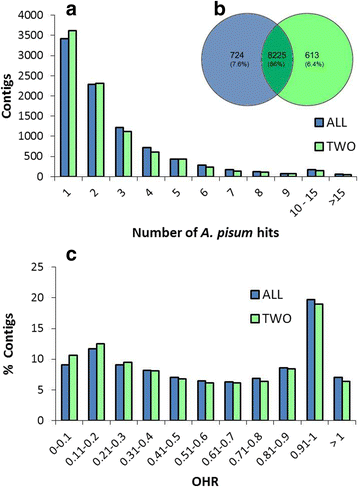
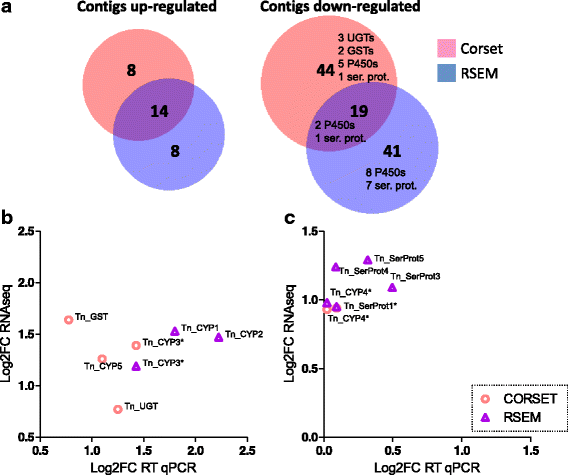
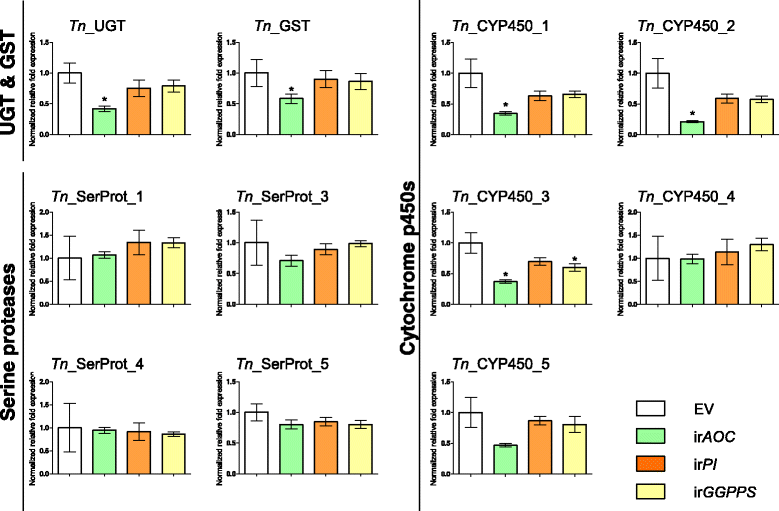
Results: We used an RNAseq approach to evaluate the global gene expression of T. notatus after feeding on a transgenic N. attenuata line which does not accumulate jasmonic acid (JA) after herbivory, and consequently accumulates very low levels of defense metabolites. Using Illumina sequencing, we generated a de novo assembled transcriptome which resulted in 63,062 contigs (putative transcript isoforms) contained in 42,610 isotigs (putative identified genes). Differential expression analysis based on RSEM-estimated transcript abundances identified 82 differentially expressed (DE) transcripts between T. notatus fed on wild-type and the defenseless plants. The same analysis conducted with Corset-estimated transcript abundances identified 59 DE clusters containing 85 transcripts. In both analyses, a larger number of DE transcripts were found down-regulated in mirids feeding on JA-silenced plants (around 70%). Among these down-regulated transcripts we identified seven transcripts possibly involved in the detoxification of N. attenuata defense metabolite, specifically, one glutathione-S-transferase (GST), one UDP-glucosyltransferase (UGT), five cytochrome P450 (P450s), and six serine proteases. Real-time quantitative PCR confirmed the down-regulation for six transcripts (encoding GST, UGT and four P450s) and revealed that their expression was only slightly decreased in mirids feeding on another N. attenuata transgenic line specifically silenced in the accumulation of diterpene glycosides, one of the many classes of JA-mediated defenses in N. attenuata.
Conclusions: The results provide a transcriptional overview of the changes in a specialist hemimetabolous insect associated with feeding on host plants depleted in chemical defenses. Overall, the analysis reveals that T. notatus responses to host plant defenses are narrow and engages P450 detoxification pathways. It further identifies candidate genes which can be tested in future experiments to understand their role in shaping the T. notatus-N. attenuata interaction.
Keywords: Coyote tobacco; Detoxification; Heteroptera; Piercing-sucking herbivory; Trophic interactions.
Figures




Similar articles
-
UVB radiation and 17-hydroxygeranyllinalool diterpene glycosides provide durable resistance against mirid (Tupiocoris notatus) attack in field-grown Nicotiana attenuata plants.Plant Cell Environ. 2013 Mar;36(3):590-606. doi: 10.1111/j.1365-3040.2012.02598.x. Epub 2012 Sep 18. Plant Cell Environ. 2013. PMID: 22897424
-
Jasmonate signaling in the field, part II: insect-guided characterization of genetic variations in jasmonate-dependent defenses of transgenic and natural Nicotiana attenuata populations.Methods Mol Biol. 2013;1011:97-109. doi: 10.1007/978-1-62703-414-2_8. Methods Mol Biol. 2013. PMID: 23615990
-
MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics.BMC Plant Biol. 2012 Nov 13;12:213. doi: 10.1186/1471-2229-12-213. BMC Plant Biol. 2012. PMID: 23148462 Free PMC article.
-
The essential role of jasmonic acid in plant-herbivore interactions--using the wild tobacco Nicotiana attenuata as a model.J Genet Genomics. 2013 Dec 20;40(12):597-606. doi: 10.1016/j.jgg.2013.10.001. Epub 2013 Nov 9. J Genet Genomics. 2013. PMID: 24377866 Review.
-
Revealing insect herbivory-induced phenolamide metabolism: from single genes to metabolic network plasticity analysis.Plant J. 2014 Aug;79(4):679-92. doi: 10.1111/tpj.12503. Epub 2014 May 21. Plant J. 2014. PMID: 24617849 Free PMC article. Review.
Cited by
-
De novo transcriptome assembly and differential gene expression analysis in different developmental stages of Agriotes sputator (click beetle).Sci Rep. 2024 Oct 18;14(1):24451. doi: 10.1038/s41598-024-74495-1. Sci Rep. 2024. PMID: 39424855 Free PMC article.
-
Multiple Known Mechanisms and a Possible Role of an Enhanced Immune System in Bt-Resistance in a Field Population of the Bollworm, Helicoverpa zea: Differences in Gene Expression with RNAseq.Int J Mol Sci. 2020 Sep 7;21(18):6528. doi: 10.3390/ijms21186528. Int J Mol Sci. 2020. PMID: 32906662 Free PMC article.
-
Midgut Transcriptional Variation of Chilo suppressalis Larvae Induced by Feeding on the Dead-End Trap Plant, Vetiveria zizanioides.Front Physiol. 2018 Aug 7;9:1067. doi: 10.3389/fphys.2018.01067. eCollection 2018. Front Physiol. 2018. PMID: 30131719 Free PMC article.
-
Genomics analysis of Drosophila sechellia response to Morinda citrifolia fruit diet.G3 (Bethesda). 2022 Sep 30;12(10):jkac153. doi: 10.1093/g3journal/jkac153. G3 (Bethesda). 2022. PMID: 35736356 Free PMC article.
-
Coagulation cascade and complement system in systemic lupus erythematosus.Oncotarget. 2017 Dec 11;9(19):14862-14881. doi: 10.18632/oncotarget.23206. eCollection 2018 Mar 13. Oncotarget. 2017. PMID: 29599912 Free PMC article.
References
-
- Ehrlich PR, Raven PH. Butterflies and plants - a study in coevolution. Evolution. 1964;18:586–608. doi: 10.2307/2406212. - DOI
-
- Heckel D. Insect detoxification and sequestration strategies. In: Voelckel C, Jander G, editors. Annual Plant Review Vol. 47 Insect-Plant Interaction. Chichester: John Wiley & Sons, Ltd; 2014. p. 77–114.
-
- Vogel H, Musser RO, Celorio-Mancera M. Transcriptome responses in herbivorous insects towards host plant and toxin feeding. In: Voelckel C, Jander G, editors. Annual Plant Review Vol. 47 Insect-Plant Interaction. Chichester: John Wiley & Sons, Ltd; 2014. p. 197–234.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

