Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing
- PMID: 27931250
- PMCID: PMC5146893
- DOI: 10.1186/s13059-016-1109-7
Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing
Abstract
Background: Single-cell micro-metastases of solid tumors often occur in the bone marrow. These disseminated tumor cells (DTCs) may resist therapy and lay dormant or progress to cause overt bone and visceral metastases. The molecular nature of DTCs remains elusive, as well as when and from where in the tumor they originate. Here, we apply single-cell sequencing to identify and trace the origin of DTCs in breast cancer.
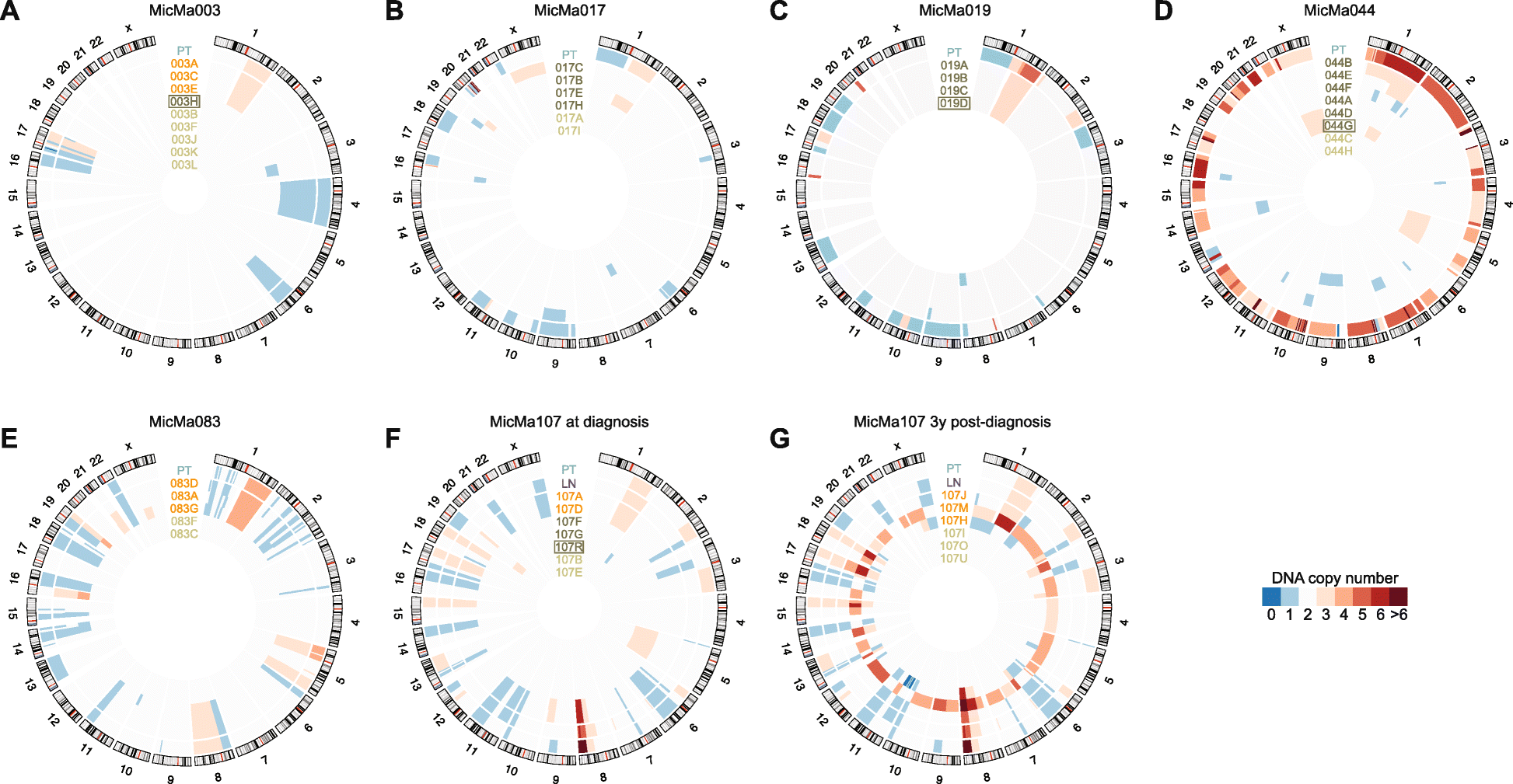
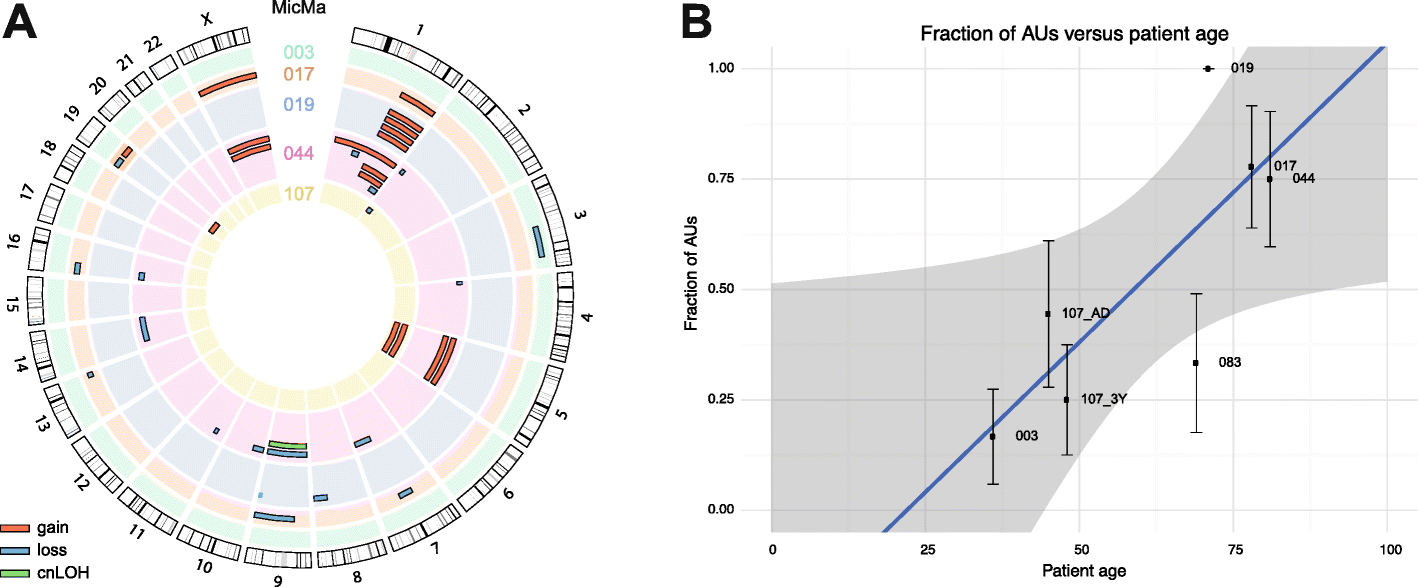
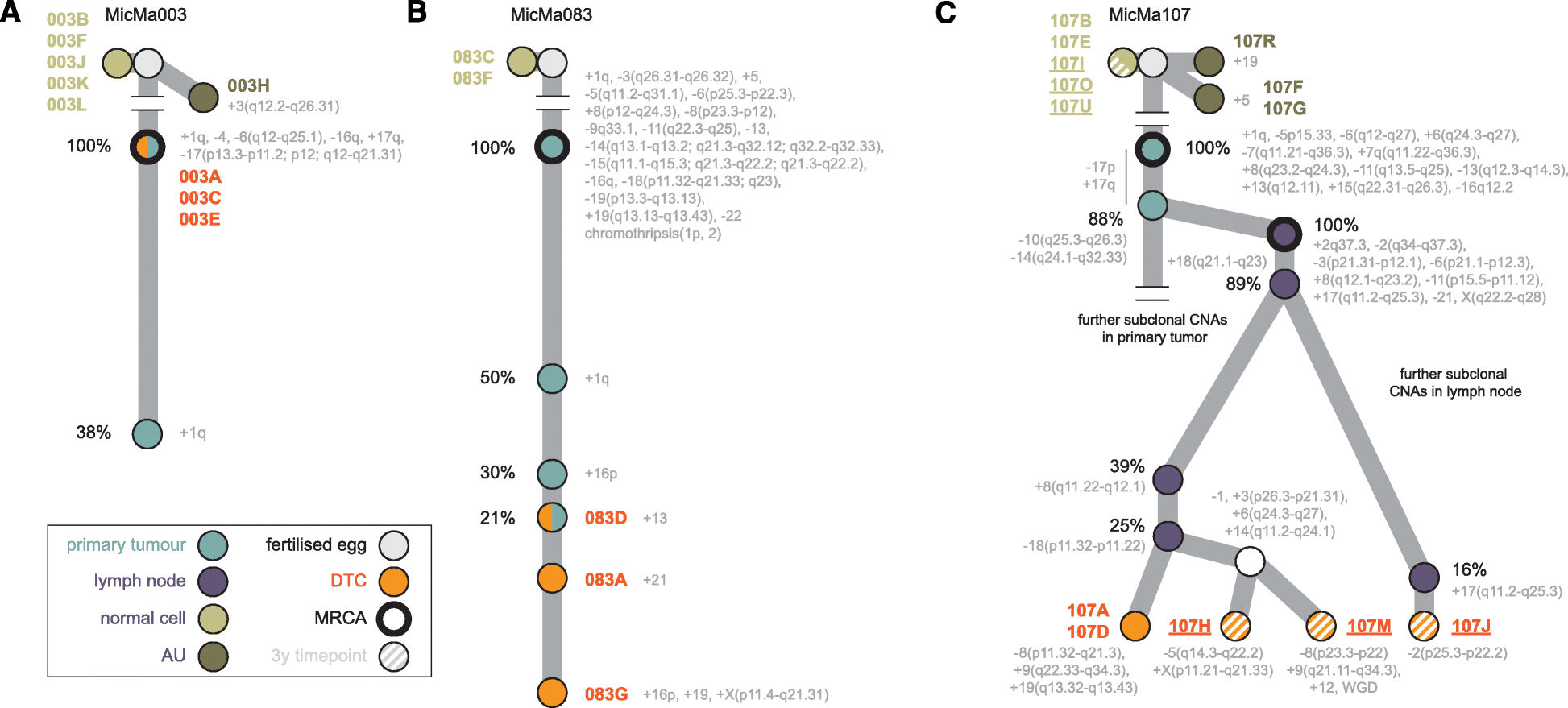
Results: We sequence the genomes of 63 single cells isolated from six non-metastatic breast cancer patients. By comparing the cells' DNA copy number aberration (CNA) landscapes with those of the primary tumors and lymph node metastasis, we establish that 53% of the single cells morphologically classified as tumor cells are DTCs disseminating from the observed tumor. The remaining cells represent either non-aberrant "normal" cells or "aberrant cells of unknown origin" that have CNA landscapes discordant from the tumor. Further analyses suggest that the prevalence of aberrant cells of unknown origin is age-dependent and that at least a subset is hematopoietic in origin. Evolutionary reconstruction analysis of bulk tumor and DTC genomes enables ordering of CNA events in molecular pseudo-time and traced the origin of the DTCs to either the main tumor clone, primary tumor subclones, or subclones in an axillary lymph node metastasis.
Conclusions: Single-cell sequencing of bone marrow epithelial-like cells, in parallel with intra-tumor genetic heterogeneity profiling from bulk DNA, is a powerful approach to identify and study DTCs, yielding insight into metastatic processes. A heterogeneous population of CNA-positive cells is present in the bone marrow of non-metastatic breast cancer patients, only part of which are derived from the observed tumor lineages.
Keywords: Disseminated tumor cells; Intra-tumor genetic heterogeneity; Metastasis; Phylogeny; Single-cell sequencing.
Figures

Comment in
-
A case of mistaken identity.Sci Transl Med. 2017 Jan 4;9(371):eaal4988. doi: 10.1126/scitranslmed.aal4988. Sci Transl Med. 2017. PMID: 28053155 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

