Population Genomics of Daphnia pulex
- PMID: 27932545
- PMCID: PMC5419477
- DOI: 10.1534/genetics.116.190611
Population Genomics of Daphnia pulex
Abstract
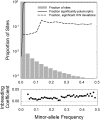
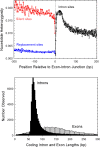
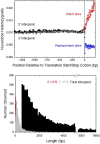
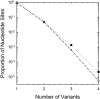
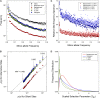
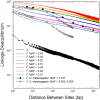
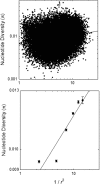
Using data from 83 isolates from a single population, the population genomics of the microcrustacean Daphnia pulex are described and compared to current knowledge for the only other well-studied invertebrate, Drosophila melanogaster These two species are quite similar with respect to effective population sizes and mutation rates, although some features of recombination appear to be different, with linkage disequilibrium being elevated at short ([Formula: see text] bp) distances in D. melanogaster and at long distances in D. pulex The study population adheres closely to the expectations under Hardy-Weinberg equilibrium, and reflects a past population history of no more than a twofold range of variation in effective population size. Fourfold redundant silent sites and a restricted region of intronic sites appear to evolve in a nearly neutral fashion, providing a powerful tool for population genetic analyses. Amino acid replacement sites are predominantly under strong purifying selection, as are a large fraction of sites in UTRs and intergenic regions, but the majority of SNPs at such sites that rise to frequencies [Formula: see text] appear to evolve in a nearly neutral fashion. All forms of genomic sites (including replacement sites within codons, and intergenic and UTR regions) appear to be experiencing an [Formula: see text] higher level of selection scaled to the power of drift in D. melanogaster, but this may in part be a consequence of recent demographic changes. These results establish D. pulex as an excellent system for future work on the evolutionary genomics of natural populations.
Keywords: Daphnia; genetic variation; linkage disequilibrium; population genomics; population size.
Copyright © 2017 by the Genetics Society of America.
Figures



References
-
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. - PubMed
-
- Ayala F. J. (Editor), 1976. Molecular Evolution, Sinauer Associates, Inc., Sunderland, MA.
-
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

