A Novel, Molybdenum-Containing Methionine Sulfoxide Reductase Supports Survival of Haemophilus influenzae in an In vivo Model of Infection
- PMID: 27933034
- PMCID: PMC5122715
- DOI: 10.3389/fmicb.2016.01743
A Novel, Molybdenum-Containing Methionine Sulfoxide Reductase Supports Survival of Haemophilus influenzae in an In vivo Model of Infection
Abstract
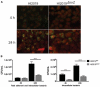
Haemophilus influenzae is a host adapted human mucosal pathogen involved in a variety of acute and chronic respiratory tract infections, including chronic obstructive pulmonary disease and asthma, all of which rely on its ability to efficiently establish continuing interactions with the host. Here we report the characterization of a novel molybdenum enzyme, TorZ/MtsZ that supports interactions of H. influenzae with host cells during growth in oxygen-limited environments. Strains lacking TorZ/MtsZ showed a reduced ability to survive in contact with epithelial cells as shown by immunofluorescence microscopy and adherence/invasion assays. This included a reduction in the ability of the strain to invade human epithelial cells, a trait that could be linked to the persistence of H. influenzae. The observation that in a murine model of H. influenzae infection, strains lacking TorZ/MtsZ were almost undetectable after 72 h of infection, while ∼3.6 × 103 CFU/mL of the wild type strain were measured under the same conditions is consistent with this view. To understand how TorZ/MtsZ mediates this effect we purified and characterized the enzyme, and were able to show that it is an S- and N-oxide reductase with a stereospecificity for S-sulfoxides. The enzyme converts two physiologically relevant sulfoxides, biotin sulfoxide and methionine sulfoxide (MetSO), with the kinetic parameters suggesting that MetSO is the natural substrate of this enzyme. TorZ/MtsZ was unable to repair sulfoxides in oxidized Calmodulin, suggesting that a role in cell metabolism/energy generation and not protein repair is the key function of this enzyme. Phylogenetic analyses showed that H. influenzae TorZ/MtsZ is only distantly related to the Escherichia coli TorZ TMAO reductase, but instead is a representative of a new, previously uncharacterized clade of molybdenum enzyme that is widely distributed within the Pasteurellaceae family of pathogenic bacteria. It is likely that MtsZ/TorZ has a similar role in supporting host/pathogen interactions in other members of the Pasteurellaceae, which includes both human and animal pathogens.
Keywords: DMSO reductase enzyme family; Haemophilus influenzae; host–pathogen interaction; methionine sulfoxide reductase; molybdenum enzymes.
Figures






References
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (2005). “Current protocols in molecular biology,” in Current Protocols in Molecular Biology ed. Janssen K. (Hoboken, NJ: John Wiley & Sons Inc; ).
-
- Barber M. J., Trimboli A., Neame P. J., Bastian N. R. (1992). Sequence similarity between dimethylsulfoxide and biotin sulfoxide reductases. FASEB J. 6 A190–A190.
LinkOut - more resources
Full Text Sources
Other Literature Sources

