Life without dUTPase
- PMID: 27933035
- PMCID: PMC5122711
- DOI: 10.3389/fmicb.2016.01768
Life without dUTPase
Abstract
Fine-tuned regulation of the cellular nucleotide pools is indispensable for faithful replication of Deoxyribonucleic Acid (DNA). The genetic information is also safeguarded by DNA damage recognition and repair processes. Uracil is one of the most frequently occurring erroneous bases in DNA; it can arise from cytosine deamination or thymine-replacing incorporation. Two enzyme activities are primarily involved in keeping DNA uracil-free: dUTPase (dUTP pyrophosphatase) activity that prevent thymine-replacing incorporation and uracil-DNA glycosylase activity that excise uracil from DNA and initiate uracil-excision repair. Both dUTPase and the most efficient uracil-DNA glycosylase (UNG) is thought to be ubiquitous in free-living organisms. In the present work, we have systematically investigated the genotype of deposited fully sequenced bacterial and Archaeal genomes. We have performed bioinformatic searches in these genomes using the already well described dUTPase and UNG gene sequences. For dUTPases, we have included the trimeric all-beta and the dimeric all-alpha families and also, the bifunctional dCTP (deoxycytidine triphosphate) deaminase-dUTPase sequences. Surprisingly, we have found that in contrast to the generally held opinion, a wide number of bacterial and Archaeal species lack all of the previously described dUTPase gene(s). The dut- genotype is present in diverse bacterial phyla indicating that loss of this (or these) gene(s) has occurred multiple times during evolution. We discuss potential survival strategies in lack of dUTPases, such as simultaneous lack or inhibition of UNG and possession of exogenous or alternate metabolic enzymes involved in uracil-DNA metabolism. The potential that genes previously not associated with dUTPase activity may still encode enzymes capable of hydrolyzing dUTP is also discussed. Our data indicate that several unicellular microorganisms may efficiently cope with a dut- genotype lacking all of the previously described dUTPase genes, and potentially leading to an unusual uracil-enrichment in their genomic DNA.
Keywords: U-DNA; UGI; Uracil-DNA-glycosylase; dUTPase; genome analysis; horizontal gene transfer; prokaryotes; uracil.
Figures
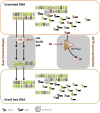

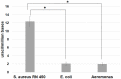
References
-
- Castillo-Acosta V. M., Aguilar-Pereyra F., Bart J. M., Navarro M., Ruiz-Perez L. M., Vidal A. E., et al. (2012a). Increased uracil insertion in DNA is cytotoxic and increases the frequency of mutation, double strand break formation and VSG switching in Trypanosoma brucei. DNA Repair (Amst) 11 986–995. 10.1016/j.dnarep.2012.09.007 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

