Comparative Genomic Analysis of the GRF Genes in Chinese Pear (Pyrus bretschneideri Rehd), Poplar (Populous), Grape (Vitis vinifera), Arabidopsis and Rice (Oryza sativa)
- PMID: 27933074
- PMCID: PMC5121280
- DOI: 10.3389/fpls.2016.01750
Comparative Genomic Analysis of the GRF Genes in Chinese Pear (Pyrus bretschneideri Rehd), Poplar (Populous), Grape (Vitis vinifera), Arabidopsis and Rice (Oryza sativa)
Abstract
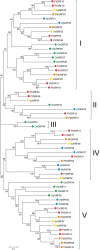
Growth-regulating factors (GRFs) are plant-specific transcription factors that have important functions in regulating plant growth and development. Previous studies on GRF family members focused either on a single or a small set of genes. Here, a comparative genomic analysis of the GRF gene family was performed in poplar (a model tree species), Arabidopsis (a model plant for annual herbaceous dicots), grape (one model plant for perennial dicots), rice (a model plant for monocots) and Chinese pear (one of the economical fruit crops). In total, 58 GRF genes were identified, 12 genes in rice (Oryza sativa), 8 genes in grape (Vitis vinifera), 9 genes in Arabidopsis thaliana, 19 genes in poplar (Populus trichocarpa) and 10 genes in Chinese pear (Pyrus bretschneideri). The GRF genes were divided into five subfamilies based on the phylogenetic analysis, which was supported by their structural analysis. Furthermore, microsynteny analysis indicated that highly conserved regions of microsynteny were identified in all of the five species tested. And Ka/Ks analysis revealed that purifying selection plays an important role in the maintenance of GRF genes. Our results provide basic information on GRF genes in five plant species and lay the foundation for future research on the functions of these genes.
Keywords: GRF; gene duplication; gene structure; microsynteny; molecular evolution.
Figures






References
-
- Baloglu M. C. (2014). Genome-wide in silico identification and comparison of Growth Regulating Factor (GRF) genes in Cucurbitaceae family. Plant Omics 7 260–270.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

