Screening and Identification of an H-2Kb-Restricted CTL Epitope within the Glycoprotein of Hantaan Virus
- PMID: 27933274
- PMCID: PMC5122572
- DOI: 10.3389/fcimb.2016.00151
Screening and Identification of an H-2Kb-Restricted CTL Epitope within the Glycoprotein of Hantaan Virus
Abstract
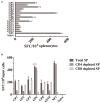
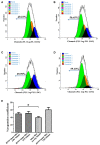
The cytotoxic T lymphocyte (CTL) response plays a key role in controlling viral infection, but only a few epitopes within the HTNV glycoprotein (GP) that are recognized by CTLs have been reported. In this study, we identified one murine HTNV GP-derived H2-Kb-restricted CTL epitope in C57BL/6 mice, which could be used to design preclinical studies of vaccines for HTNV infection. First, 15 8-mer peptides were selected from the HTNV GP amino acid sequence based on a percentile rank of <=1% by IEDB which is the most comprehensive collection of epitope prediction and analysis tool. A lower percentile rank indicates higher affinity and higher immune response. In the case of the consensus method, we also evaluated the binding score of peptide-binding affinity by the BIMAS software to confirm that all peptides were able to bind H2-Kb. Second, one novel GP-derived CTL epitope, GP6 aa456-aa463 (ITSLFSLL), was identified in the splenocytes of HTNV-infected mice using the IFN-γ ELISPOT assay. Third, a single peptide vaccine was administered to C57BL/6 mice to evaluate the immunogenic potential of the identified peptides. ELISPOT and cell-mediated cytotoxicity assays showed that this peptide vaccine induced a strong IFN-γ response and potent cytotoxicity in immunized mice. Last, we demonstrated that the peptide-vaccinated mice had partial protection from challenge with HTNV. In conclusion, we identified an H2-Kb-restricted CTL epitope with involvement in the host immune response to HTNV infection.
Keywords: HTNV; IFN-γ; epitope; glycoprotein; infection.
Figures





Similar articles
-
Novel Identified HLA-A*0201-Restricted Hantaan Virus Glycoprotein Cytotoxic T-Cell Epitopes Could Effectively Induce Protective Responses in HLA-A2.1/Kb Transgenic Mice May Associate with the Severity of Hemorrhagic Fever with Renal Syndrome.Front Immunol. 2017 Dec 12;8:1797. doi: 10.3389/fimmu.2017.01797. eCollection 2017. Front Immunol. 2017. PMID: 29312318 Free PMC article.
-
Design and synthesis of HLA-A*02-restricted Hantaan virus multiple-antigenic peptide for CD8+ T cells.Virol J. 2020 Jan 31;17(1):15. doi: 10.1186/s12985-020-1290-x. Virol J. 2020. PMID: 32005266 Free PMC article.
-
Protective CD8+ T-cell response against Hantaan virus infection induced by immunization with designed linear multi-epitope peptides in HLA-A2.1/Kb transgenic mice.Virol J. 2020 Oct 7;17(1):146. doi: 10.1186/s12985-020-01421-y. Virol J. 2020. PMID: 33028368 Free PMC article.
-
Identification of an H2-Kb or H2-Db restricted and glypican-3-derived cytotoxic T-lymphocyte epitope peptide.Int J Oncol. 2013 Mar;42(3):831-8. doi: 10.3892/ijo.2013.1793. Epub 2013 Jan 23. Int J Oncol. 2013. PMID: 23354275 Free PMC article.
-
Single-chain lipopeptide vaccines for the induction of virus-specific cytotoxic T cell responses in randomly selected populations.Mol Immunol. 2001 Dec;38(6):423-31. doi: 10.1016/s0161-5890(01)00078-5. Mol Immunol. 2001. PMID: 11741692 Review.
Cited by
-
Integrative Analysis of HTNV Glycoprotein Derived MHC II Epitopes by In Silico Prediction and Experimental Validation.Front Cell Infect Microbiol. 2021 Jul 19;11:671694. doi: 10.3389/fcimb.2021.671694. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34350130 Free PMC article.
-
Novel Identified HLA-A*0201-Restricted Hantaan Virus Glycoprotein Cytotoxic T-Cell Epitopes Could Effectively Induce Protective Responses in HLA-A2.1/Kb Transgenic Mice May Associate with the Severity of Hemorrhagic Fever with Renal Syndrome.Front Immunol. 2017 Dec 12;8:1797. doi: 10.3389/fimmu.2017.01797. eCollection 2017. Front Immunol. 2017. PMID: 29312318 Free PMC article.
-
Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2Kb Offer New Options To Combat Cryptosporidiosis.mBio. 2023 Feb 28;14(1):e0266622. doi: 10.1128/mbio.02666-22. Epub 2023 Jan 5. mBio. 2023. PMID: 36602309 Free PMC article.
-
The Adaptive Immune Response against Bunyavirales.Viruses. 2024 Mar 21;16(3):483. doi: 10.3390/v16030483. Viruses. 2024. PMID: 38543848 Free PMC article. Review.
-
Design and synthesis of HLA-A*02-restricted Hantaan virus multiple-antigenic peptide for CD8+ T cells.Virol J. 2020 Jan 31;17(1):15. doi: 10.1186/s12985-020-1290-x. Virol J. 2020. PMID: 32005266 Free PMC article.
References
-
- Ennis F. A., Cruz J., Spiropoulou C. F., Waite D., Peters C. J., Nichol S. T., et al. . (1997). Hantavirus pulmonary syndrome: CD8+ and CD4+ cytotoxic T lymphocytes to epitopes on Sin Nombre virus nucleocapsid protein isolated during acute illness. Virology 238, 380–390. 10.1006/viro.1997.8827 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

