Development of Correlative Cryo-soft X-ray Tomography and Stochastic Reconstruction Microscopy. A Study of Cholesterol Crystal Early Formation in Cells
- PMID: 27934213
- PMCID: PMC6530486
- DOI: 10.1021/jacs.6b07584
Development of Correlative Cryo-soft X-ray Tomography and Stochastic Reconstruction Microscopy. A Study of Cholesterol Crystal Early Formation in Cells
Abstract
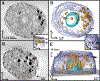
We have developed a high resolution correlative method involving cryo-soft X-ray tomography (cryo-SXT) and stochastic optical reconstruction microscopy (STORM), which provides information in three dimensions on large cellular volumes at 70 nm resolution. Cryo-SXT morphologically identified and localized aggregations of carbon-rich materials. STORM identified specific markers on the desired epitopes, enabling colocalization between the identified objects, in this case cholesterol crystals, and the cellular environment. The samples were studied under ambient and cryogenic conditions without dehydration or heavy metal staining. The early events of cholesterol crystal development were investigated in relation to atherosclerosis, using as model macrophage cell cultures enriched with LDL particles. Atherosclerotic plaques build up in arteries in a slow process involving cholesterol crystal accumulation. Cholesterol crystal deposition is a crucial stage in the pathological cascade. Our results show that cholesterol crystals can be identified and imaged at a very early stage on the cell plasma membrane and in intracellular locations. This technique can in principle be applied to other biological samples where specific molecular identification is required in conjunction with high resolution 3D-imaging.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





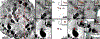
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

