The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences
- PMID: 27934697
- PMCID: PMC5131817
- DOI: 10.1101/gr.204255.116
The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences
Abstract
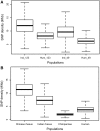
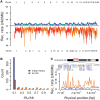
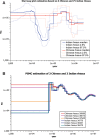
Rhesus macaques (Macaca mulatta) are the most widely used nonhuman primate in biomedical research, have the largest natural geographic distribution of any nonhuman primate, and have been the focus of much evolutionary and behavioral investigation. Consequently, rhesus macaques are one of the most thoroughly studied nonhuman primate species. However, little is known about genome-wide genetic variation in this species. A detailed understanding of extant genomic variation among rhesus macaques has implications for the use of this species as a model for studies of human health and disease, as well as for evolutionary population genomics. Whole-genome sequencing analysis of 133 rhesus macaques revealed more than 43.7 million single-nucleotide variants, including thousands predicted to alter protein sequences, transcript splicing, and transcription factor binding sites. Rhesus macaques exhibit 2.5-fold higher overall nucleotide diversity and slightly elevated putative functional variation compared with humans. This functional variation in macaques provides opportunities for analyses of coding and noncoding variation, and its cellular consequences. Despite modestly higher levels of nonsynonymous variation in the macaques, the estimated distribution of fitness effects and the ratio of nonsynonymous to synonymous variants suggest that purifying selection has had stronger effects in rhesus macaques than in humans. Demographic reconstructions indicate this species has experienced a consistently large but fluctuating population size. Overall, the results presented here provide new insights into the population genomics of nonhuman primates and expand genomic information directly relevant to primate models of human disease.
© 2016 Xue et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Abegg C, Thierry B. 2002. Macaque evolution and dispersal in insular south-east Asia. Biol J Linnean Soc 75: 555–576.
-
- Allentoft ME, Sikora M, Sjögren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522: 167–172. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
