Extensive translational regulation during seed germination revealed by polysomal profiling
- PMID: 27935038
- PMCID: PMC5347915
- DOI: 10.1111/nph.14355
Extensive translational regulation during seed germination revealed by polysomal profiling
Abstract
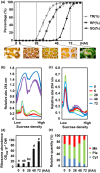
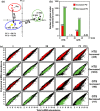
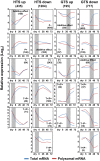
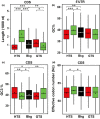
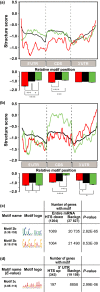
This work investigates the extent of translational regulation during seed germination. The polysome occupancy of each gene is determined by genome-wide profiling of total mRNA and polysome-associated mRNA. This reveals extensive translational regulation during Arabidopsis thaliana seed germination. The polysome occupancy of thousands of individual mRNAs changes to a large extent during the germination process. Intriguingly, these changes are restricted to two temporal phases (shifts) during germination, seed hydration and germination. Sequence features, such as upstream open reading frame number, transcript length, mRNA stability, secondary structures, and the presence and location of specific motifs correlated with this translational regulation. These features differed significantly between the two shifts, indicating that independent mechanisms regulate translation during seed germination. This study reveals substantial translational dynamics during seed germination and identifies development-dependent sequence features and cis elements that correlate with the translation control, uncovering a novel and important layer of gene regulation during seed germination.
Keywords: Arabidopsis; RNA structure; germination; imbibition; polysomal profiling; ribosome; seedling establishment; translatomics.
© 2016 The Authors. New Phytologist © 2016 New Phytologist Trust.
Figures

References
-
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch‐Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. 2008. Genome‐scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

