Probing bacterial cell biology using image cytometry
- PMID: 27935200
- PMCID: PMC5663501
- DOI: 10.1111/mmi.13591
Probing bacterial cell biology using image cytometry
Abstract
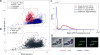
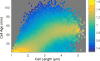
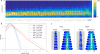
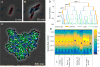
Advances in automated fluorescence microscopy have made snapshot and time-lapse imaging of bacterial cells commonplace, yet fundamental challenges remain in analysis. The vast quantity of data collected in high-throughput experiments requires a fast and reliable automated method to analyze fluorescence intensity and localization, cell morphology and proliferation as well as other descriptors. Inspired by effective yet tractable methods of population-level analysis using flow cytometry, we have developed a framework and tools for facilitating analogous analyses in image cytometry. These tools can both visualize and gate (generate subpopulations) more than 70 cell descriptors, including cell size, age and fluorescence. The method is well suited to multi-well imaging, analysis of bacterial cultures with high cell density (thousands of cells per frame) and complete cell cycle imaging. We give a brief description of the analysis of four distinct applications to emphasize the broad applicability of the tool.
© 2016 John Wiley & Sons Ltd.
Conflict of interest statement
No conflict of interest declared.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

