Identification of the fitness determinants of budding yeast on a natural substrate
- PMID: 27935595
- PMCID: PMC5364353
- DOI: 10.1038/ismej.2016.170
Identification of the fitness determinants of budding yeast on a natural substrate
Abstract
The budding yeasts are prime models in genomics and cell biology, but the ecological factors that determine their success in non-human-associated habitats is poorly understood. In North America Saccharomyces yeasts are present on the bark of deciduous trees, where they feed on bark and sap exudates. In the North East, Saccharomyces paradoxus is found on maples, which makes maple sap a natural substrate for this species. We measured growth rates of S. paradoxus natural isolates on maple sap and found variation along a geographical gradient not explained by the inherent variation observed under optimal laboratory conditions. We used a functional genomic screen to reveal the ecologically relevant genes and conditions required for optimal growth in this substrate. We found that the allantoin degradation pathway is required for optimal growth in maple sap, in particular genes necessary for allantoate utilization, which we demonstrate is the major nitrogen source available to yeast in this environment. Growth with allantoin or allantoate as the sole nitrogen source recapitulated the variation in growth rates in maple sap among strains. We also show that two lineages of S. paradoxus display different life-history traits on allantoin and allantoate media, highlighting the ecological relevance of this pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
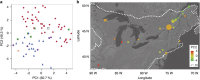



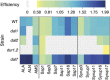
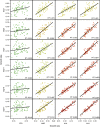

Similar articles
-
Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics.FEMS Yeast Res. 2002 Jan;1(4):299-306. doi: 10.1111/j.1567-1364.2002.tb00048.x. FEMS Yeast Res. 2002. PMID: 12702333
-
The interaction of Saccharomyces paradoxus with its natural competitors on oak bark.Mol Ecol. 2015 Apr;24(7):1596-610. doi: 10.1111/mec.13120. Epub 2015 Mar 23. Mol Ecol. 2015. PMID: 25706044 Free PMC article.
-
Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Saccharomyces paradoxus in North America.FEMS Yeast Res. 2014 Mar;14(2):281-8. doi: 10.1111/1567-1364.12100. Epub 2013 Oct 25. FEMS Yeast Res. 2014. PMID: 24119009
-
The rise of yeast population genomics.C R Biol. 2011 Aug-Sep;334(8-9):612-9. doi: 10.1016/j.crvi.2011.05.009. Epub 2011 Jul 1. C R Biol. 2011. PMID: 21819942 Review.
-
Ecological and evolutionary genomics of Saccharomyces cerevisiae.Mol Ecol. 2006 Mar;15(3):575-91. doi: 10.1111/j.1365-294X.2006.02778.x. Mol Ecol. 2006. PMID: 16499686 Review.
Cited by
-
Competition experiments in a soil microcosm reveal the impact of genetic and biotic factors on natural yeast populations.ISME J. 2020 Jun;14(6):1410-1421. doi: 10.1038/s41396-020-0612-8. Epub 2020 Feb 20. ISME J. 2020. PMID: 32080356 Free PMC article.
-
Evolutionary biology through the lens of budding yeast comparative genomics.Nat Rev Genet. 2017 Oct;18(10):581-598. doi: 10.1038/nrg.2017.49. Epub 2017 Jul 17. Nat Rev Genet. 2017. PMID: 28714481 Review.
-
Natural trait variation across Saccharomycotina species.FEMS Yeast Res. 2024 Jan 9;24:foae002. doi: 10.1093/femsyr/foae002. FEMS Yeast Res. 2024. PMID: 38218591 Free PMC article. Review.
-
Systematic Evaluation of Biotic and Abiotic Factors in Antifungal Microorganism Screening.Microorganisms. 2024 Jul 10;12(7):1396. doi: 10.3390/microorganisms12071396. Microorganisms. 2024. PMID: 39065164 Free PMC article.
-
Persistence of Resident and Transplanted Genotypes of the Undomesticated Yeast Saccharomyces paradoxus in Forest Soil.mSphere. 2018 Jun 20;3(3):e00211-18. doi: 10.1128/mSphere.00211-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29925673 Free PMC article.
References
-
- Atkins C. (1982). Ureide metabolism and the significance of ureides in legumes. InSubba Rao NSed Advances in agricultural microbiology Elsevier: Amsterdam, Netherlands, pp 53–88.
-
- Barnes RL. (1959). Formation of allantoin and allantoic acid from adenine in leaves of Acer saccharinum L. Nature 184: 1944–1944. - PubMed
-
- Barnes RL. (1963). Organic nitrogen compounds in tree xylem sap. Forest Sci 9: 98–102.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

