Patient-Specific Bacteroides Genome Variants in Pouchitis
- PMID: 27935837
- PMCID: PMC5111406
- DOI: 10.1128/mBio.01713-16
Patient-Specific Bacteroides Genome Variants in Pouchitis
Abstract
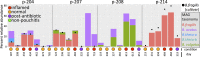
A 2-year longitudinal microbiome study of 22 patients who underwent colectomy with an ileal pouch anal anastomosis detected significant increases in distinct populations of Bacteroides during 9 of 11 patient visits that coincided with inflammation (pouchitis). Oligotyping and metagenomic short-read annotation identified Bacteroides populations that occurred in early samples, bloomed during inflammation, and reappeared after antibiotic treatment. Targeted cultivation of Bacteroides isolates from the same individual at multiple time points and from several patients detected subtle genomic changes, including the identification of rapidly evolving genomic elements that differentiate isogenic strains of Bacteroides fragilis from the mucosa versus lumen. Each patient harbored Bacteroides spp. that are closely related to commonly occurring clinical isolates, including Bacteroides ovatus, B. thetaiotaomicron, B. vulgatus, and B. fragilis, which contained unique loci in different patients for synthesis of capsular polysaccharides. The presence of unique Bacteroides capsular polysaccharide loci within different hosts and between the lumen and mucosa may represent adaptations to stimulate, suppress, and evade host-specific immune responses at different microsites of the ileal pouch.
Importance: This longitudinal study provides an opportunity to describe shifts in the microbiomes of individual patients who suffer from ulcerative colitis (UC) prior to and following inflammation. Pouchitis serves as a model for UC with a predictable incidence of disease onset and enables prospective longitudinal investigations of UC etiology prior to inflammation. Because of insufficient criteria for predicting which patients will develop UC or pouchitis, the interpretation of cross-sectional study designs suffers from lack of information about the microbiome structure and host gene expression patterns that directly correlate with the onset of disease. Our unique longitudinal study design allows each patient to serve as their own control, providing information about the state of the microbiome and host prior to and during the course of disease. Of significance to the broader community, this study identifies microbial strains that may have genetic elements that trigger the onset of disease in susceptible hosts.
Copyright © 2016 Vineis et al.
Figures



References
-
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, et al. . 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. doi:10.1038/nature11582. - DOI - PMC - PubMed
-
- Paziewska A, Horbacka K, Goryca K, Mikula M, Jarosz D, Dabrowska M, Krokowicz P, Karon J, Ostrowski J. 2015. Transcriptional changes between uninflamed ulcerative colitis and familial adenomatous polyposis pouch mucosa can be attributed to an altered immune response. Acta Biochim Pol 62:69–75. doi:10.18388/abp.2014_778. - DOI - PubMed
-
- Young VB, Raffals LH, Huse SM, Vital M, Dai D, Schloss PD, Brulc JM, Antonopoulos DA, Arrieta RL, Kwon JH, Reddy KG, Hubert NA, Grim SL, Vineis JH, Dalal S, Morrison HG, Eren AM, Meyer F, Schmidt TM, Tiedje JM, Chang EB, Sogin ML. 2013. Multiphasic analysis of the temporal development of the distal gut microbiota in patients following ileal pouch anal anastomosis. Microbiome 1:9. doi:10.1186/2049-2618-1-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
