Splicing factor gene mutations in hematologic malignancies
- PMID: 27940478
- PMCID: PMC5345729
- DOI: 10.1182/blood-2016-10-692400
Splicing factor gene mutations in hematologic malignancies
Abstract
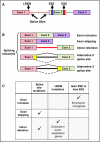
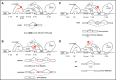
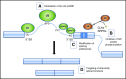
Alternative splicing generates a diversity of messenger RNA (mRNA) transcripts from a single mRNA precursor and contributes to the complexity of our proteome. Splicing is perturbed by a variety of mechanisms in cancer. Recurrent mutations in splicing factors have emerged as a hallmark of several hematologic malignancies. Splicing factor mutations tend to occur in the founding clone of myeloid cancers, and these mutations have recently been identified in blood cells from normal, healthy elderly individuals with clonal hematopoiesis who are at increased risk of subsequently developing a hematopoietic malignancy, suggesting that these mutations contribute to disease initiation. Splicing factor mutations change the pattern of splicing in primary patient and mouse hematopoietic cells and alter hematopoietic differentiation and maturation in animal models. Recent developments in this field are reviewed here, with an emphasis on the clinical consequences of splicing factor mutations, mechanistic insights from animal models, and implications for development of novel therapies targeting the precursor mRNA splicing pathway.
© 2017 by The American Society of Hematology.
Figures
Similar articles
-
RNA splicing factors in normal hematopoiesis and hematologic malignancies: novel therapeutic targets and strategies.J Leukoc Biol. 2023 Feb 1;113(2):149-163. doi: 10.1093/jleuko/qiac015. J Leukoc Biol. 2023. PMID: 36822179
-
Splicing factor mutations in hematologic malignancies.Blood. 2021 Aug 26;138(8):599-612. doi: 10.1182/blood.2019004260. Blood. 2021. PMID: 34157091 Free PMC article. Review.
-
Splicing Factor Mutations in Cancer.Adv Exp Med Biol. 2016;907:215-28. doi: 10.1007/978-3-319-29073-7_9. Adv Exp Med Biol. 2016. PMID: 27256388
-
Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy.Immunol Rev. 2015 Jan;263(1):257-78. doi: 10.1111/imr.12241. Immunol Rev. 2015. PMID: 25510282 Review.
-
Mutations of RNA splicing factors in hematological malignancies.Cancer Lett. 2017 Nov 28;409:1-8. doi: 10.1016/j.canlet.2017.08.042. Epub 2017 Sep 6. Cancer Lett. 2017. PMID: 28888996 Review.
Cited by
-
Cancer synthetic vulnerabilities to protein arginine methyltransferase inhibitors.Curr Opin Pharmacol. 2021 Aug;59:33-42. doi: 10.1016/j.coph.2021.04.004. Epub 2021 May 27. Curr Opin Pharmacol. 2021. PMID: 34052526 Free PMC article. Review.
-
U2AF1 in various neoplastic diseases and relevant targeted therapies for malignant cancers with complex mutations (Review).Oncol Rep. 2024 Jan;51(1):5. doi: 10.3892/or.2023.8664. Epub 2023 Nov 17. Oncol Rep. 2024. PMID: 37975232 Free PMC article. Review.
-
Genomic landscape of neutrophilic leukemias of ambiguous diagnosis.Blood. 2019 Sep 12;134(11):867-879. doi: 10.1182/blood.2019000611. Epub 2019 Jul 31. Blood. 2019. PMID: 31366621 Free PMC article.
-
The impact of spliceosome mutations in MDS.Hemasphere. 2019 Jun 30;3(Suppl):132-134. doi: 10.1097/HS9.0000000000000218. eCollection 2019 Jun. Hemasphere. 2019. PMID: 35309818 Free PMC article. No abstract available.
-
Transcription Factors, R-Loops and Deubiquitinating Enzymes: Emerging Targets in Myelodysplastic Syndromes and Acute Myeloid Leukemia.Cancers (Basel). 2021 Jul 26;13(15):3753. doi: 10.3390/cancers13153753. Cancers (Basel). 2021. PMID: 34359655 Free PMC article. Review.
References
-
- Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. - PubMed
-
- Sakharkar MK, Chow VT, Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol. 2004;4(4):387-393. - PubMed
-
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749-761. - PubMed
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413-1415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

