Contribution of Pentose Catabolism to Molecular Hydrogen Formation by Targeted Disruption of Arabinose Isomerase (araA) in the Hyperthermophilic Bacterium Thermotoga maritima
- PMID: 27940539
- PMCID: PMC5288831
- DOI: 10.1128/AEM.02631-16
Contribution of Pentose Catabolism to Molecular Hydrogen Formation by Targeted Disruption of Arabinose Isomerase (araA) in the Hyperthermophilic Bacterium Thermotoga maritima
Abstract
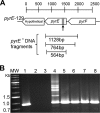
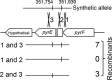
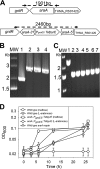
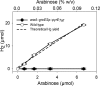
Thermotoga maritima ferments a broad range of sugars to form acetate, carbon dioxide, traces of lactate, and near theoretic yields of molecular hydrogen (H2). In this organism, the catabolism of pentose sugars such as arabinose depends on the interaction of the pentose phosphate pathway with the Embden-Myerhoff and Entner-Doudoroff pathways. Although the values for H2 yield have been determined using pentose-supplemented complex medium and predicted by metabolic pathway reconstruction, the actual effect of pathway elimination on hydrogen production has not been reported due to the lack of a genetic method for the creation of targeted mutations. Here, a spontaneous and genetically stable pyrE deletion mutant was isolated and used as a recipient to refine transformation methods for its repair by homologous recombination. To verify the occurrence of recombination and to assess the frequency of crossover events flanking the deleted region, a synthetic pyrE allele, encoding synonymous nucleotide substitutions, was used. Targeted inactivation of araA (encoding arabinose isomerase) in the pyrE mutant was accomplished using a divergent, codon-optimized Thermosipho africanus pyrE allele fused to the T. maritima groES promoter as a genetic marker. Mutants lacking araA were unable to catabolize arabinose in a defined medium. The araA mutation was then repaired using targeted recombination. Levels of synthesis of H2 using arabinose-supplemented complex medium by wild-type and araA mutant cell lines were compared. The difference between strains provided a direct measurement of H2 production that was dependent on arabinose consumption. Development of a targeted recombination system for genetic manipulation of T. maritima provides a new strategy to explore H2 formation and life at an extremely high temperature in the bacterial domain.
Importance: We describe here the development of a genetic system for manipulation of Thermotoga maritima T. maritima is a hyperthermophilic anaerobic bacterium that is well known for its efficient synthesis of molecular hydrogen (H2) from the fermentation of sugars. Despite considerable efforts to advance compatible genetic methods, chromosome manipulation has remained elusive and hindered use of T. maritima or its close relatives as model hyperthermophiles. Lack of a genetic method also prevented efforts to manipulate specific metabolic pathways to measure their contributions to H2 yield. To overcome this barrier, a homologous chromosomal recombination method was developed and used to characterize the contribution of arabinose catabolism to H2 formation. We report here a stable genetic method for a hyperthermophilic bacterium that will advance studies on the basic and synthetic biology of Thermotogales.
Keywords: anaerobes; biohydrogen; extremophiles; genetic systems; homologous recombination.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Huber R, Langworthy TA, Konig H, Thomm M, Woese CR, Sleytr UB, Stetter KO. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol 144:324–333.
-
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329. doi:10.1038/20601. - DOI - PubMed
-
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT, Nelson KE, Nesbø CL, Doolittle WF, Gogarten JP, Noll KM. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci U S A 106:5865–5870. doi:10.1073/pnas.0901260106. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

