Kinetic isotope effects reveal early transition state of protein lysine methyltransferase SET8
- PMID: 27940912
- PMCID: PMC5206543
- DOI: 10.1073/pnas.1609032114
Kinetic isotope effects reveal early transition state of protein lysine methyltransferase SET8
Abstract
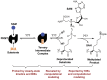
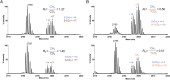
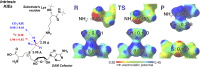
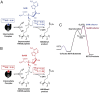
Protein lysine methyltransferases (PKMTs) catalyze the methylation of protein substrates, and their dysregulation has been linked to many diseases, including cancer. Accumulated evidence suggests that the reaction path of PKMT-catalyzed methylation consists of the formation of a cofactor(cosubstrate)-PKMT-substrate complex, lysine deprotonation through dynamic water channels, and a nucleophilic substitution (SN2) transition state for transmethylation. However, the molecular characters of the proposed process remain to be elucidated experimentally. Here we developed a matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) method and corresponding mathematic matrix to determine precisely the ratios of isotopically methylated peptides. This approach may be generally applicable for examining the kinetic isotope effects (KIEs) of posttranslational modifying enzymes. Protein lysine methyltransferase SET8 is the sole PKMT to monomethylate histone 4 lysine 20 (H4K20) and its function has been implicated in normal cell cycle progression and cancer metastasis. We therefore implemented the MS-based method to measure KIEs and binding isotope effects (BIEs) of the cofactor S-adenosyl-l-methionine (SAM) for SET8-catalyzed H4K20 monomethylation. A primary intrinsic 13C KIE of 1.04, an inverse intrinsic α-secondary CD3 KIE of 0.90, and a small but statistically significant inverse CD3 BIE of 0.96, in combination with computational modeling, revealed that SET8-catalyzed methylation proceeds through an early, asymmetrical SN2 transition state with the C-N and C-S distances of 2.35-2.40 Å and 2.00-2.05 Å, respectively. This transition state is further supported by the KIEs, BIEs, and steady-state kinetics with the SAM analog Se-adenosyl-l-selenomethionine (SeAM) as a cofactor surrogate. The distinct transition states between protein methyltransferases present the opportunity to design selective transition-state analog inhibitors.
Keywords: BIE; KIE; PKMT; PMT; methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

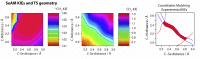
 , 25 μM;
, 25 μM;  , 50 μM; and ∆, 100 μM). The initial linear range varies with cofactor and substrate concentrations. (B and C) Lineweaver–Burk analysis of the initial velocities vs. varied concentrations of the H4K20 substrate (●, 30 μM; ■, 20 μM; ▲, 15 μM; ▼, 10 μM; and ◆, 8 μM) and SAM (●, 6 μM; ▼,15 μM; ◆, 25 μM;
, 50 μM; and ∆, 100 μM). The initial linear range varies with cofactor and substrate concentrations. (B and C) Lineweaver–Burk analysis of the initial velocities vs. varied concentrations of the H4K20 substrate (●, 30 μM; ■, 20 μM; ▲, 15 μM; ▼, 10 μM; and ◆, 8 μM) and SAM (●, 6 μM; ▼,15 μM; ◆, 25 μM;  , 50 μM; and
, 50 μM; and  , 100 μM). All of the initial rates converge in the secondary quadrant. (D) Global fit of the initial velocities to a general, bisubstrate kinetic mechanism against SAM concentration to afford kcat, Km,SAM, Km,H4K20 and α according to Eqs. S1 and S2 with kcat = 7.0 ± 0.8 min−1, Km,peptide = 40 ± 8 µM, Km,cofactor = 16 ± 6 µM, and α = 1.4 ± 0.8 (45).
, 100 μM). All of the initial rates converge in the secondary quadrant. (D) Global fit of the initial velocities to a general, bisubstrate kinetic mechanism against SAM concentration to afford kcat, Km,SAM, Km,H4K20 and α according to Eqs. S1 and S2 with kcat = 7.0 ± 0.8 min−1, Km,peptide = 40 ± 8 µM, Km,cofactor = 16 ± 6 µM, and α = 1.4 ± 0.8 (45).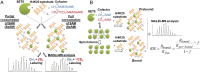


Similar articles
-
Product specificity and mechanism of protein lysine methyltransferases: insights from the histone lysine methyltransferase SET8.Biochemistry. 2008 Jun 24;47(25):6671-7. doi: 10.1021/bi800244s. Biochemistry. 2008. PMID: 18512960
-
Substrate-Differentiated Transition States of SET7/9-Catalyzed Lysine Methylation.J Am Chem Soc. 2019 May 22;141(20):8064-8067. doi: 10.1021/jacs.9b02553. Epub 2019 May 14. J Am Chem Soc. 2019. PMID: 31034218 Free PMC article.
-
Transition state for the NSD2-catalyzed methylation of histone H3 lysine 36.Proc Natl Acad Sci U S A. 2016 Feb 2;113(5):1197-201. doi: 10.1073/pnas.1521036113. Epub 2016 Jan 19. Proc Natl Acad Sci U S A. 2016. PMID: 26787850 Free PMC article.
-
Approaching the catalytic mechanism of protein lysine methyltransferases by biochemical and simulation techniques.Crit Rev Biochem Mol Biol. 2024 Feb-Apr;59(1-2):20-68. doi: 10.1080/10409238.2024.2318547. Epub 2024 Mar 7. Crit Rev Biochem Mol Biol. 2024. PMID: 38449437 Review.
-
Protein Lysine Methyltransferases Inhibitors.Curr Med Chem. 2023;30(27):3060-3089. doi: 10.2174/0929867329666220829151257. Curr Med Chem. 2023. PMID: 36043747 Review.
Cited by
-
Crystallographic and Computational Characterization of Methyl Tetrel Bonding in S-Adenosylmethionine-Dependent Methyltransferases.Molecules. 2018 Nov 13;23(11):2965. doi: 10.3390/molecules23112965. Molecules. 2018. PMID: 30428636 Free PMC article.
-
Trimethyllysine: From Carnitine Biosynthesis to Epigenetics.Int J Mol Sci. 2020 Dec 11;21(24):9451. doi: 10.3390/ijms21249451. Int J Mol Sci. 2020. PMID: 33322546 Free PMC article. Review.
-
The transition to magic bullets - transition state analogue drug design.Medchemcomm. 2018 Aug 28;9(12):1983-1993. doi: 10.1039/c8md00372f. eCollection 2018 Dec 1. Medchemcomm. 2018. PMID: 30627387 Free PMC article. Review.
-
Stuffed Methyltransferase Catalyzes the Penultimate Step of Pyochelin Biosynthesis.Biochemistry. 2019 Feb 12;58(6):665-678. doi: 10.1021/acs.biochem.8b00716. Epub 2018 Dec 27. Biochemistry. 2019. PMID: 30525512 Free PMC article.
-
Chemical and Biochemical Perspectives of Protein Lysine Methylation.Chem Rev. 2018 Jul 25;118(14):6656-6705. doi: 10.1021/acs.chemrev.8b00008. Epub 2018 Jun 21. Chem Rev. 2018. PMID: 29927582 Free PMC article. Review.
References
-
- Cleland WW. The use of isotope effects to determine enzyme mechanisms. Arch Biochem Biophys. 2005;433(1):2–12. - PubMed
-
- Cleland WW. Isotope effects: Determination of enzyme transition state structure. Methods Enzymol. 1995;249:341–373. - PubMed
-
- Świderek K, Paneth P. Binding isotope effects. Chem Rev. 2013;113(10):7851–7879. - PubMed
-
- Northrop DB. Steady-state analysis of kinetic isotope effects in enzymic reactions. Biochemistry. 1975;14(12):2644–2651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

