Computational design of a homotrimeric metalloprotein with a trisbipyridyl core
- PMID: 27940918
- PMCID: PMC5206526
- DOI: 10.1073/pnas.1600188113
Computational design of a homotrimeric metalloprotein with a trisbipyridyl core
Abstract
Metal-chelating heteroaryl small molecules have found widespread use as building blocks for coordination-driven, self-assembling nanostructures. The metal-chelating noncanonical amino acid (2,2'-bipyridin-5yl)alanine (Bpy-ala) could, in principle, be used to nucleate specific metalloprotein assemblies if introduced into proteins such that one assembly had much lower free energy than all alternatives. Here we describe the use of the Rosetta computational methodology to design a self-assembling homotrimeric protein with [Fe(Bpy-ala)3]2+ complexes at the interface between monomers. X-ray crystallographic analysis of the homotrimer showed that the design process had near-atomic-level accuracy: The all-atom rmsd between the design model and crystal structure for the residues at the protein interface is ∼1.4 Å. These results demonstrate that computational protein design together with genetically encoded noncanonical amino acids can be used to drive formation of precisely specified metal-mediated protein assemblies that could find use in a wide range of photophysical applications.
Keywords: computational protein design; metalloproteins; noncanonical amino acids; protein self-assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



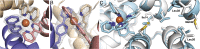



Similar articles
-
Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy.J Am Chem Soc. 2013 Sep 11;135(36):13393-9. doi: 10.1021/ja403503m. Epub 2013 Aug 29. J Am Chem Soc. 2013. PMID: 23924187 Free PMC article.
-
Metal-chelating non-canonical amino acids in metalloprotein engineering and design.Curr Opin Struct Biol. 2018 Aug;51:170-176. doi: 10.1016/j.sbi.2018.06.001. Epub 2018 Jul 3. Curr Opin Struct Biol. 2018. PMID: 29980106 Review.
-
Computational approaches for de novo design and redesign of metal-binding sites on proteins.Biosci Rep. 2017 Mar 27;37(2):BSR20160179. doi: 10.1042/BSR20160179. Print 2017 Apr 28. Biosci Rep. 2017. PMID: 28167677 Free PMC article. Review.
-
Design and engineering of metalloproteins containing unnatural amino acids or non-native metal-containing cofactors.Curr Opin Chem Biol. 2005 Apr;9(2):118-26. doi: 10.1016/j.cbpa.2005.02.017. Curr Opin Chem Biol. 2005. PMID: 15811795 Review.
-
Metal-Directed Design of Supramolecular Protein Assemblies.Methods Enzymol. 2016;580:223-50. doi: 10.1016/bs.mie.2016.05.009. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27586336 Free PMC article.
Cited by
-
Noncanonical Amino Acids: Bringing New-to-Nature Functionalities to Biocatalysis.Chem Rev. 2024 Oct 9;124(19):10877-10923. doi: 10.1021/acs.chemrev.4c00136. Epub 2024 Sep 27. Chem Rev. 2024. PMID: 39329413 Free PMC article. Review.
-
A Hydroxyquinoline-Based Unnatural Amino Acid for the Design of Novel Artificial Metalloenzymes.Chembiochem. 2020 Nov 2;21(21):3077-3081. doi: 10.1002/cbic.202000306. Epub 2020 Jul 17. Chembiochem. 2020. PMID: 32585070 Free PMC article.
-
Structural Insights into How Protein Environments Tune the Spectroscopic Properties of a Noncanonical Amino Acid Fluorophore.Biochemistry. 2020 Sep 22;59(37):3401-3410. doi: 10.1021/acs.biochem.0c00474. Epub 2020 Sep 3. Biochemistry. 2020. PMID: 32845612 Free PMC article.
-
Diverse protein assembly driven by metal and chelating amino acids with selectivity and tunability.Nat Commun. 2019 Dec 5;10(1):5545. doi: 10.1038/s41467-019-13491-w. Nat Commun. 2019. PMID: 31804480 Free PMC article.
-
Better together: building protein oligomers naturally and by design.Biochem Soc Trans. 2019 Dec 20;47(6):1773-1780. doi: 10.1042/BST20190283. Biochem Soc Trans. 2019. PMID: 31803901 Free PMC article. Review.
References
-
- Kaes C, Katz A, Hosseini MW. Bipyridine: The most widely used ligand. A review of molecules comprising at least two 2,2′-bipyridine units. Chem Rev. 2000;100(10):3553–3590. - PubMed
-
- Leininger S, Olenyuk B, Stang PJ. Self-assembly of discrete cyclic nanostructures mediated by transition metals. Chem Rev. 2000;100(3):853–908. - PubMed
-
- Lieberman M, Sasaki T. Iron(II) organizes a synthetic peptide into three-helix bundles. J Am Chem Soc. 1991;113(4):1470–1471.
-
- Ghadiri MR, Soares C, Choi C. A convergent approach to protein design. Metal ion-assisted spontaneous self-assembly of a polypeptide into a triple-helix bundle protein. J Am Chem Soc. 1992;114(3):825–831.
-
- Xie J, Liu W, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angew Chem Int Ed Engl. 2007;46(48):9239–9242. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

