A Multivariate Mixture Model to Estimate the Accuracy of Glycosaminoglycan Identifications Made by Tandem Mass Spectrometry (MS/MS) and Database Search
- PMID: 27941081
- PMCID: PMC5294212
- DOI: 10.1074/mcp.M116.062588
A Multivariate Mixture Model to Estimate the Accuracy of Glycosaminoglycan Identifications Made by Tandem Mass Spectrometry (MS/MS) and Database Search
Abstract
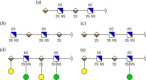
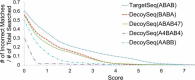
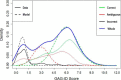
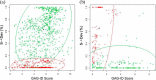
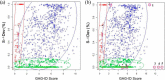
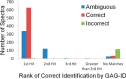
We present a statistical model to estimate the accuracy of derivatized heparin and heparan sulfate (HS) glycosaminoglycan (GAG) assignments to tandem mass (MS/MS) spectra made by the first published database search application, GAG-ID. Employing a multivariate expectation-maximization algorithm, this statistical model distinguishes correct from ambiguous and incorrect database search results when computing the probability that heparin/HS GAG assignments to spectra are correct based upon database search scores. Using GAG-ID search results for spectra generated from a defined mixture of 21 synthesized tetrasaccharide sequences as well as seven spectra of longer defined oligosaccharides, we demonstrate that the computed probabilities are accurate and have high power to discriminate between correctly, ambiguously, and incorrectly assigned heparin/HS GAGs. This analysis makes it possible to filter large MS/MS database search results with predictable false identification error rates.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

