Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode
- PMID: 27941644
- PMCID: PMC5192498
- DOI: 10.3390/genes7120122
Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode
Abstract
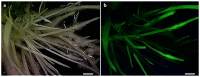
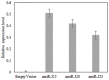
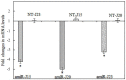
The soybean cyst nematode (SCN), Heterodera glycines, is one of the most important pests limiting soybean production worldwide. Novel approaches to managing this pest have focused on gene silencing of target nematode sequences using RNA interference (RNAi). With the discovery of endogenous microRNAs as a mode of gene regulation in plants, artificial microRNA (amiRNA) methods have become an alternative method for gene silencing, with the advantage that they can lead to more specific silencing of target genes than traditional RNAi vectors. To explore the application of amiRNAs for improving soybean resistance to SCN, three nematode genes (designated as J15, J20, and J23) were targeted using amiRNA vectors. The transgenic soybean hairy roots, transformed independently with these three amiRNA vectors, showed significant reductions in SCN population densities in bioassays. Expression of the targeted genes within SCN eggs were downregulated in populations feeding on transgenic hairy roots. Our results provide evidence that host-derived amiRNA methods have great potential to improve soybean resistance to SCN. This approach should also limit undesirable phenotypes associated with off-target effects, which is an important consideration for commercialization of transgenic crops.
Keywords: artificial microRNA; resistance; soybean; soybean cyst nematode.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures





Similar articles
-
Host-derived gene silencing of parasite fitness genes improves resistance to soybean cyst nematodes in stable transgenic soybean.Theor Appl Genet. 2019 Sep;132(9):2651-2662. doi: 10.1007/s00122-019-03379-0. Epub 2019 Jun 22. Theor Appl Genet. 2019. PMID: 31230117 Free PMC article.
-
A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance.BMC Plant Biol. 2010 Jun 9;10:104. doi: 10.1186/1471-2229-10-104. BMC Plant Biol. 2010. PMID: 20529370 Free PMC article.
-
Enhanced resistance to soybean cyst nematode in transgenic soybean via host-induced silencing of vital Heterodera glycines genes.Transgenic Res. 2022 Apr;31(2):239-248. doi: 10.1007/s11248-022-00298-7. Epub 2022 Feb 8. Transgenic Res. 2022. PMID: 35133563
-
Molecular Basis of Soybean Resistance to Soybean Aphids and Soybean Cyst Nematodes.Plants (Basel). 2019 Sep 26;8(10):374. doi: 10.3390/plants8100374. Plants (Basel). 2019. PMID: 31561499 Free PMC article. Review.
-
Soybean cyst nematode detection and management: a review.Plant Methods. 2022 Sep 7;18(1):110. doi: 10.1186/s13007-022-00933-8. Plant Methods. 2022. PMID: 36071455 Free PMC article. Review.
Cited by
-
Engineered Artificial MicroRNA Precursors Facilitate Cloning and Gene Silencing in Arabidopsis and Rice.Int J Mol Sci. 2019 Nov 10;20(22):5620. doi: 10.3390/ijms20225620. Int J Mol Sci. 2019. PMID: 31717686 Free PMC article.
-
Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing.BMC Genomics. 2017 Aug 2;18(1):572. doi: 10.1186/s12864-017-3963-4. BMC Genomics. 2017. PMID: 28768484 Free PMC article.
-
Small RNA-based plant protection against diseases.Front Plant Sci. 2022 Aug 18;13:951097. doi: 10.3389/fpls.2022.951097. eCollection 2022. Front Plant Sci. 2022. PMID: 36061762 Free PMC article. Review.
-
Host-derived gene silencing of parasite fitness genes improves resistance to soybean cyst nematodes in stable transgenic soybean.Theor Appl Genet. 2019 Sep;132(9):2651-2662. doi: 10.1007/s00122-019-03379-0. Epub 2019 Jun 22. Theor Appl Genet. 2019. PMID: 31230117 Free PMC article.
-
Species-specific microRNA discovery and target prediction in the soybean cyst nematode.Sci Rep. 2023 Oct 17;13(1):17657. doi: 10.1038/s41598-023-44469-w. Sci Rep. 2023. PMID: 37848601 Free PMC article.
References
-
- Koenning S.R., Wrather J.A. Suppression of soybean yield potential in the continental united states from plant diseases estimated from 2006 to 2009. Plant Health Prog. 2010 doi: 10.1094/PHP-2010-1122-01-RS. - DOI
-
- Davis E.L., Tylka G.L. Soybean cyst nematode disease. Plant Health Instr. 2000 doi: 10.1094/PHI-I-2000-0725-01. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

