Anaphase B
- PMID: 27941648
- PMCID: PMC5192431
- DOI: 10.3390/biology5040051
Anaphase B
Abstract
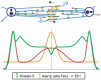
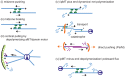
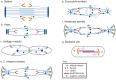
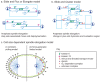
Anaphase B spindle elongation is characterized by the sliding apart of overlapping antiparallel interpolar (ip) microtubules (MTs) as the two opposite spindle poles separate, pulling along disjoined sister chromatids, thereby contributing to chromosome segregation and the propagation of all cellular life. The major biochemical "modules" that cooperate to mediate pole-pole separation include: (i) midzone pushing or (ii) braking by MT crosslinkers, such as kinesin-5 motors, which facilitate or restrict the outward sliding of antiparallel interpolar MTs (ipMTs); (iii) cortical pulling by disassembling astral MTs (aMTs) and/or dynein motors that pull aMTs outwards; (iv) ipMT plus end dynamics, notably net polymerization; and (v) ipMT minus end depolymerization manifest as poleward flux. The differential combination of these modules in different cell types produces diversity in the anaphase B mechanism. Combinations of antagonist modules can create a force balance that maintains the dynamic pre-anaphase B spindle at constant length. Tipping such a force balance at anaphase B onset can initiate and control the rate of spindle elongation. The activities of the basic motor filament components of the anaphase B machinery are controlled by a network of non-motor MT-associated proteins (MAPs), for example the key MT cross-linker, Ase1p/PRC1, and various cell-cycle kinases, phosphatases, and proteases. This review focuses on the molecular mechanisms of anaphase B spindle elongation in eukaryotic cells and briefly mentions bacterial DNA segregation systems that operate by spindle elongation.
Keywords: anaphase B; mitotic motors; poleward flux; spindle elongation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

