ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice
- PMID: 27941798
- PMCID: PMC5285448
- DOI: 10.1038/ng.3744
ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice
Abstract
Genes encoding subunits of SWI/SNF (BAF) chromatin-remodeling complexes are collectively mutated in ∼20% of all human cancers. Although ARID1A is the most frequent target of mutations, the mechanism by which its inactivation promotes tumorigenesis is unclear. Here we demonstrate that Arid1a functions as a tumor suppressor in the mouse colon, but not the small intestine, and that invasive ARID1A-deficient adenocarcinomas resemble human colorectal cancer (CRC). These tumors lack deregulation of APC/β-catenin signaling components, which are crucial gatekeepers in common forms of intestinal cancer. We find that ARID1A normally targets SWI/SNF complexes to enhancers, where they function in coordination with transcription factors to facilitate gene activation. ARID1B preserves SWI/SNF function in ARID1A-deficient cells, but defects in SWI/SNF targeting and control of enhancer activity cause extensive dysregulation of gene expression. These findings represent an advance in colon cancer modeling and implicate enhancer-mediated gene regulation as a principal tumor-suppressor function of ARID1A.
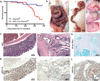
Figures





References
-
- Garraway LA, Lander ES. Lessons from the Cancer Genome. Cell. 2013;153(1):17–37. http://doi.org/10.1016/j.cell.2013.03.002. - DOI - PubMed
-
- Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. - PubMed
-
- Hamilton SR, Bosman FT, Boffetta P, et al. Carcinoma of the colon and rectum. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. Lyon: IARC Press; 2010. pp. 134–146.
-
- Marjou el F, Janssen K-P, Chang BH-J, Li M, Hindie V, Chan L, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis (New York, N.Y. : 2000) 2004;39(3):186–193. http://doi.org/10.1002/gene.20042. - DOI - PubMed
Methods-only References
-
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6656–6661. http://doi.org/10.1073/pnas.0801802105. - DOI - PMC - PubMed
-
- Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. The Journal of Biological Chemistry. 1973;248(7):2536–2541. - PubMed
-
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997v2 [q-bio.GN] 2013
-
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research. 2012;22(3):568–576. http://doi.org/10.1101/gr.129684.111. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

