Methyltransferase-Directed Labeling of Biomolecules and its Applications
- PMID: 27943567
- PMCID: PMC5502580
- DOI: 10.1002/anie.201608625
Methyltransferase-Directed Labeling of Biomolecules and its Applications
Abstract
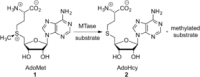
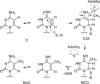
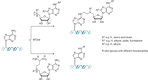
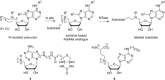
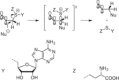
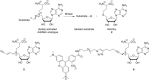
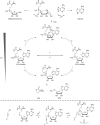
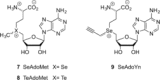
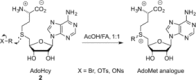
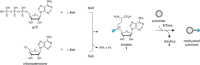
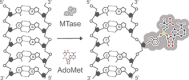
Methyltransferases (MTases) form a large family of enzymes that methylate a diverse set of targets, ranging from the three major biopolymers to small molecules. Most of these MTases use the cofactor S-adenosyl-l-Methionine (AdoMet) as a methyl source. In recent years, there have been significant efforts toward the development of AdoMet analogues with the aim of transferring moieties other than simple methyl groups. Two major classes of AdoMet analogues currently exist: doubly-activated molecules and aziridine based molecules, each of which employs a different approach to achieve transalkylation rather than transmethylation. In this review, we discuss the various strategies for labelling and functionalizing biomolecules using AdoMet-dependent MTases and AdoMet analogues. We cover the synthetic routes to AdoMet analogues, their stability in biological environments and their application in transalkylation reactions. Finally, some perspectives are presented for the potential use of AdoMet analogues in biology research, (epi)genetics and nanotechnology.
Keywords: DNA functionalization; S-adenosyl methionine; methyltransferases; protein modification; transalkylation.
© 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
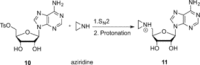



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

