Quantitative proteomic profiling of paired cancerous and normal colon epithelial cells isolated freshly from colorectal cancer patients
- PMID: 27943637
- PMCID: PMC5418098
- DOI: 10.1002/prca.201600155
Quantitative proteomic profiling of paired cancerous and normal colon epithelial cells isolated freshly from colorectal cancer patients
Abstract
Purpose: The heterogeneous structure in tumor tissues from colorectal cancer (CRC) patients excludes an informative comparison between tumors and adjacent normal tissues. Here, we develop and apply a strategy to compare paired cancerous (CEC) versus normal (NEC) epithelial cells enriched from patients and discover potential biomarkers and therapeutic targets for CRC.
Experimental design: CEC and NEC cells are respectively isolated from five different tumor and normal locations in the resected colon tissue from each patient (N = 12 patients) using an optimized epithelial cell adhesion molecule (EpCAM)-based enrichment approach. An ion current-based quantitative method is employed to perform comparative proteomic analysis for each patient.
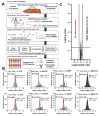
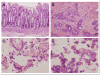
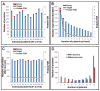
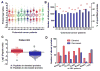
Results: A total of 458 altered proteins that are common among >75% of patients are observed and selected for further investigation. Besides known findings such as deregulation of mitochondrial function, tricarboxylic acid cycle, and RNA post-transcriptional modification, functional analysis further revealed RAN signaling pathway, small nucleolar ribonucleoproteins (snoRNPs), and infection by RNA viruses are altered in CEC cells. A selection of the altered proteins of interest is validated by immunohistochemistry analyses.
Conclusion and clinical relevance: The informative comparison between matched CEC and NEC enhances our understanding of molecular mechanisms of CRC development and provides biomarker candidates and new pathways for therapeutic intervention.
Keywords: Biomarker discovery; Colorectal cancer; Ion current-based analysis; Tumor cell enrichment.
© 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


Similar articles
-
Wnt-driven LARGE2 mediates laminin-adhesive O-glycosylation in human colonic epithelial cells and colorectal cancer.Cell Commun Signal. 2020 Jun 25;18(1):102. doi: 10.1186/s12964-020-00561-6. Cell Commun Signal. 2020. PMID: 32586342 Free PMC article.
-
Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis.J Proteomics. 2013 Dec 6;94:473-85. doi: 10.1016/j.jprot.2013.10.020. Epub 2013 Oct 24. J Proteomics. 2013. PMID: 24332065 Clinical Trial.
-
Interplay between post-translational cyclooxygenase-2 modifications and the metabolic and proteomic profile in a colorectal cancer cohort.World J Gastroenterol. 2019 Jan 28;25(4):433-446. doi: 10.3748/wjg.v25.i4.433. World J Gastroenterol. 2019. PMID: 30700940 Free PMC article.
-
EphA3 promotes malignant transformation of colorectal epithelial cells by upregulating oncogenic pathways.Cancer Lett. 2016 Dec 28;383(2):195-203. doi: 10.1016/j.canlet.2016.10.004. Epub 2016 Oct 6. Cancer Lett. 2016. PMID: 27721017
-
Lamin A: a putative colonic epithelial stem cell biomarker which identifies colorectal tumours with a more aggressive phenotype.Biochem Soc Trans. 2008 Dec;36(Pt 6):1350-3. doi: 10.1042/BST0361350. Biochem Soc Trans. 2008. PMID: 19021554 Review.
Cited by
-
Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives.Cancers (Basel). 2022 Nov 11;14(22):5545. doi: 10.3390/cancers14225545. Cancers (Basel). 2022. PMID: 36428637 Free PMC article. Review.
-
High-quality and robust protein quantification in large clinical/pharmaceutical cohorts with IonStar proteomics investigation.Nat Protoc. 2023 Mar;18(3):700-731. doi: 10.1038/s41596-022-00780-w. Epub 2022 Dec 9. Nat Protoc. 2023. PMID: 36494494 Free PMC article. Review.
-
Proteomic Profiles and Biological Processes of Relapsed vs. Non-Relapsed Pediatric Hodgkin Lymphoma.Int J Mol Sci. 2020 Mar 22;21(6):2185. doi: 10.3390/ijms21062185. Int J Mol Sci. 2020. PMID: 32235718 Free PMC article. Clinical Trial.
-
MS1 ion current-based quantitative proteomics: A promising solution for reliable analysis of large biological cohorts.Mass Spectrom Rev. 2019 Nov;38(6):461-482. doi: 10.1002/mas.21595. Epub 2019 Mar 28. Mass Spectrom Rev. 2019. PMID: 30920002 Free PMC article. Review.
References
-
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. - PubMed
-
- Seeff LC, Royalty J, Helsel WE, Kammerer WG, et al. Clinical outcomes from the CDC’s Colorectal Cancer Screening Demonstration Program. Cancer. 2013;119(Suppl 15):2820–2833. - PubMed
-
- Bretthauer M, Hoff G. Prevention and early diagnosis of colorectal cancer. Tidsskrift for den Norske laegeforening : tidsskrift for praktisk medicin, ny raekke. 2007;127:2688–2691. - PubMed
-
- O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. Journal of the National Cancer Institute. 2004;96:1420–1425. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

