NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer
- PMID: 27951586
- PMCID: PMC5468516
- DOI: 10.1038/nature20597
NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer
Abstract
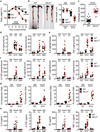
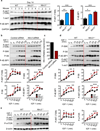
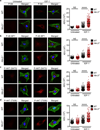
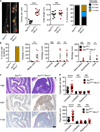
NLRs (nucleotide-binding domain and leucine-rich repeats) belong to a large family of cytoplasmic sensors that regulate an extraordinarily diverse range of biological functions. One of these functions is to contribute to immunity against infectious diseases, but dysregulation of their functional activity leads to the development of inflammatory and autoimmune diseases. Cytoplasmic innate immune sensors, including NLRs, are central regulators of intestinal homeostasis. NLRC3 (also known as CLR16.2 or NOD3) is a poorly characterized member of the NLR family and was identified in a genomic screen for genes encoding proteins bearing leucine-rich repeats (LRRs) and nucleotide-binding domains. Expression of NLRC3 is drastically reduced in the tumour tissue of patients with colorectal cancer compared to healthy tissues, highlighting an undefined potential function for this sensor in the development of cancer. Here we show that mice lacking NLRC3 are hyper-susceptible to colitis and colorectal tumorigenesis. The effect of NLRC3 is most dominant in enterocytes, in which it suppresses activation of the mTOR signalling pathways and inhibits cellular proliferation and stem-cell-derived organoid formation. NLRC3 associates with PI3Ks and blocks activation of the PI3K-dependent kinase AKT following binding of growth factor receptors or Toll-like receptor 4. These findings reveal a key role for NLRC3 as an inhibitor of the mTOR pathways, mediating protection against colorectal cancer.
Figures





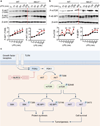
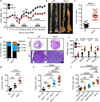


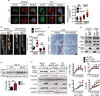
Comment in
-
Tumour immunology: NLRC3 inhibits mTOR in colorectal cancer.Nat Rev Immunol. 2017 Feb;17(2):79. doi: 10.1038/nri.2016.152. Epub 2017 Jan 3. Nat Rev Immunol. 2017. PMID: 28044056 No abstract available.
-
Colorectal cancer: A new innate immune sensor - functions from inside the colonic epithelium.Nat Rev Gastroenterol Hepatol. 2017 Apr;14(4):199-200. doi: 10.1038/nrgastro.2017.10. Epub 2017 Feb 15. Nat Rev Gastroenterol Hepatol. 2017. PMID: 28196986 No abstract available.
-
NLR members in inflammation-associated carcinogenesis.Cell Mol Immunol. 2017 May;14(5):403-405. doi: 10.1038/cmi.2017.14. Epub 2017 Apr 3. Cell Mol Immunol. 2017. PMID: 28366939 Free PMC article. No abstract available.
References
-
- Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nature reviews. Immunology. 2008;8:372–379. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

