Genome-Wide CRISPR Screen Identifies Regulators of Mitogen-Activated Protein Kinase as Suppressors of Liver Tumors in Mice
- PMID: 27956228
- PMCID: PMC6204228
- DOI: 10.1053/j.gastro.2016.12.002
Genome-Wide CRISPR Screen Identifies Regulators of Mitogen-Activated Protein Kinase as Suppressors of Liver Tumors in Mice
Abstract
Background & aims: It has been a challenge to identify liver tumor suppressors or oncogenes due to the genetic heterogeneity of these tumors. We performed a genome-wide screen to identify suppressors of liver tumor formation in mice, using CRISPR-mediated genome editing.
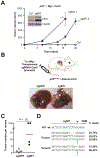
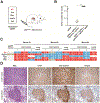
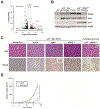
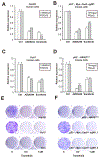
Methods: We performed a genome-wide CRISPR/Cas9-based knockout screen of P53-null mouse embryonic liver progenitor cells that overexpressed MYC. We infected p53-/-;Myc;Cas9 hepatocytes with the mGeCKOa lentiviral library of 67,000 single-guide RNAs (sgRNAs), targeting 20,611 mouse genes, and transplanted the transduced cells subcutaneously into nude mice. Within 1 month, all the mice that received the sgRNA library developed subcutaneous tumors. We performed high-throughput sequencing of tumor DNA and identified sgRNAs increased at least 8-fold compared to the initial cell pool. To validate the top 10 candidate tumor suppressors from this screen, we collected data from patients with hepatocellular carcinoma (HCC) using the Cancer Genome Atlas and COSMIC databases. We used CRISPR to inactivate candidate tumor suppressor genes in p53-/-;Myc;Cas9 cells and transplanted them subcutaneously into nude mice; tumor formation was monitored and tumors were analyzed by histology and immunohistochemistry. Mice with liver-specific disruption of p53 were given hydrodynamic tail-vein injections of plasmids encoding Myc and sgRNA/Cas9 designed to disrupt candidate tumor suppressors; growth of tumors and metastases was monitored. We compared gene expression profiles of liver cells with vs without tumor suppressor gene disrupted by sgRNA/Cas9. Genes found to be up-regulated after tumor suppressor loss were examined in liver cancer cell lines; their expression was knocked down using small hairpin RNAs, and tumor growth was examined in nude mice. Effects of the MEK inhibitors AZD6244, U0126, and trametinib, or the multi-kinase inhibitor sorafenib, were examined in human and mouse HCC cell lines.
Results: We identified 4 candidate liver tumor suppressor genes not previously associated with liver cancer (Nf1, Plxnb1, Flrt2, and B9d1). CRISPR-mediated knockout of Nf1, a negative regulator of RAS, accelerated liver tumor formation in mice. Loss of Nf1 or activation of RAS up-regulated the liver progenitor cell markers HMGA2 and SOX9. RAS pathway inhibitors suppressed the activation of the Hmga2 and Sox9 genes that resulted from loss of Nf1 or oncogenic activation of RAS. Knockdown of HMGA2 delayed formation of xenograft tumors from cells that expressed oncogenic RAS. In human HCCs, low levels of NF1 messenger RNA or high levels of HMGA2 messenger RNA were associated with shorter patient survival time. Liver cancer cells with inactivation of Plxnb1, Flrt2, and B9d1 formed more tumors in mice and had increased levels of mitogen-activated protein kinase phosphorylation.
Conclusions: Using a CRISPR-based strategy, we identified Nf1, Plxnb1, Flrt2, and B9d1 as suppressors of liver tumor formation. We validated the observation that RAS signaling, via mitogen-activated protein kinase, contributes to formation of liver tumors in mice. We associated decreased levels of NF1 and increased levels of its downstream protein HMGA2 with survival times of patients with HCC. Strategies to inhibit or reduce HMGA2 might be developed to treat patients with liver cancer.
Keywords: CRISPR Screen; Liver Cancer; Mouse Model; Tumor Suppressor Genes.
Copyright © 2017 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures



Comment in
-
Cancer Gene Discovery in Hepatocellular Carcinoma: The CRISPR/CAS9 Accelerator.Gastroenterology. 2017 Apr;152(5):941-943. doi: 10.1053/j.gastro.2017.02.031. Epub 2017 Mar 1. Gastroenterology. 2017. PMID: 28259794 No abstract available.
References
-
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226–1239. - PubMed
-
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674–87. - PubMed
-
- Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015;15:653–67. - PubMed
-
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians 2015;65:87–108. - PubMed
-
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

