Inventory of Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae in France as Assessed by a Multicenter Study
- PMID: 27956424
- PMCID: PMC5328551
- DOI: 10.1128/AAC.01911-16
Inventory of Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae in France as Assessed by a Multicenter Study
Abstract
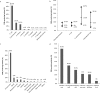
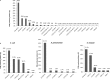
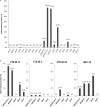
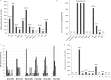
The objective of this study was to perform an inventory of the extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae isolates responsible for infections in French hospitals and to assess the mechanisms associated with ESBL diffusion. A total of 200 nonredundant ESBL-producing Enterobacteriaceae strains isolated from clinical samples were collected during a multicenter study performed in 18 representative French hospitals. Antibiotic resistance genes were identified by PCR and sequencing experiments. The clonal relatedness between isolates was investigated by the use of the DiversiLab system. ESBL-encoding plasmids were compared by PCR-based replicon typing and plasmid multilocus sequence typing. CTX-M-15, CTX-M-1, CTX-M-14, and SHV-12 were the most prevalent ESBLs (8% to 46.5%). The three CTX-M-type EBSLs were significantly observed in Escherichia coli (37.1%, 24.2%, and 21.8%, respectively), and CTX-M-15 was the predominant ESBL in Klebsiella pneumoniae (81.1%). SHV-12 was associated with ESBL-encoding Enterobacter cloacae strains (37.9%). qnrB, aac(6')-Ib-cr, and aac(3)-II genes were the main plasmid-mediated resistance genes, with prevalences ranging between 19.5% and 45% according to the ESBL results. Molecular typing did not identify wide clonal diffusion. Plasmid analysis suggested the diffusion of low numbers of ESBL-encoding plasmids, especially in K. pneumoniae and E. cloacae However, the ESBL-encoding genes were observed in different plasmid replicons according to the bacterial species. The prevalences of ESBL subtypes differ according to the Enterobacteriaceae species. Plasmid spread is a key determinant of this epidemiology, and the link observed between the ESBL-encoding plasmids and the bacterial host explains the differences observed in the Enterobacteriaceae species.
Keywords: AAC(3)-II; AAC(6′)-Ib; ESBL; plasmid; plasmid-mediated quinolone resistance; plasmid-mediated resistance.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 59:165–174. - PubMed
-
- Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

