Inhibiting translation elongation can aid genome duplication in Escherichia coli
- PMID: 27956500
- PMCID: PMC5389703
- DOI: 10.1093/nar/gkw1254
Inhibiting translation elongation can aid genome duplication in Escherichia coli
Abstract
Conflicts between replication and transcription challenge chromosome duplication. Escherichia coli replisome movement along transcribed DNA is promoted by Rep and UvrD accessory helicases with Δrep ΔuvrD cells being inviable under rapid growth conditions. We have discovered that mutations in a tRNA gene, aspT, in an aminoacyl tRNA synthetase, AspRS, and in a translation factor needed for efficient proline-proline bond formation, EF-P, suppress Δrep ΔuvrD lethality. Thus replication-transcription conflicts can be alleviated by the partial sacrifice of a mechanism that reduces replicative barriers, namely translating ribosomes that reduce RNA polymerase backtracking. Suppression depends on RelA-directed synthesis of (p)ppGpp, a signalling molecule that reduces replication-transcription conflicts, with RelA activation requiring ribosomal pausing. Levels of (p)ppGpp in these suppressors also correlate inversely with the need for Rho activity, an RNA translocase that can bind to emerging transcripts and displace transcription complexes. These data illustrate the fine balance between different mechanisms in facilitating gene expression and genome duplication and demonstrate that accessory helicases are a major determinant of this balance. This balance is also critical for other aspects of bacterial survival: the mutations identified here increase persistence indicating that similar mutations could arise in naturally occurring bacterial populations facing antibiotic challenge.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

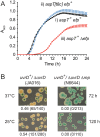


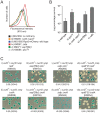
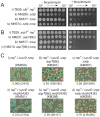

References
-
- McGlynn P., Savery N.J., Dillingham M.S.. The conflict between DNA replication and transcription. Mol. Microbiol. 2012; 85:12–20. - PubMed
-
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006; 34:1062–1066. - PubMed
-
- Komissarova N., Kashlev M.. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997; 272:15329–15338. - PubMed
MeSH terms
Substances
Grants and funding
- BB/J014826/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0800970/MRC_/Medical Research Council/United Kingdom
- BB/I001859/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I001859/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I003142/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

