Biasing genome-editing events toward precise length deletions with an RNA-guided TevCas9 dual nuclease
- PMID: 27956611
- PMCID: PMC5206545
- DOI: 10.1073/pnas.1616343114
Biasing genome-editing events toward precise length deletions with an RNA-guided TevCas9 dual nuclease
Abstract
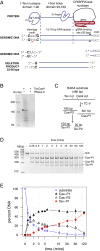
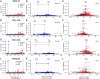
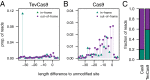
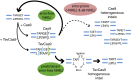
The CRISPR/Cas9 nuclease is commonly used to make gene knockouts. The blunt DNA ends generated by cleavage can be efficiently ligated by the classical nonhomologous end-joining repair pathway (c-NHEJ), regenerating the target site. This repair creates a cycle of cleavage, ligation, and target site regeneration that persists until sufficient modification of the DNA break by alternative NHEJ prevents further Cas9 cutting, generating a heterogeneous population of insertions and deletions typical of gene knockouts. Here, we develop a strategy to escape this cycle and bias events toward defined length deletions by creating an RNA-guided dual active site nuclease that generates two noncompatible DNA breaks at a target site, effectively deleting the majority of the target site such that it cannot be regenerated. The TevCas9 nuclease, a fusion of the I-TevI nuclease domain to Cas9, functions robustly in HEK293 cells and generates 33- to 36-bp deletions at frequencies up to 40%. Deep sequencing revealed minimal processing of TevCas9 products, consistent with protection of the DNA ends from exonucleolytic degradation and repair by the c-NHEJ pathway. Directed evolution experiments identified I-TevI variants with broadened targeting range, making TevCas9 an easy-to-use reagent. Our results highlight how the sequence-tolerant cleavage properties of the I-TevI homing endonuclease can be harnessed to enhance Cas9 applications, circumventing the cleavage and ligation cycle and biasing genome-editing events toward defined length deletions.
Keywords: CRISPR/Cas9; I-TevI homing endonuclease; NHEJ; genome editing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. - PubMed
-
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. - PubMed
-
- Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

