A single splice site mutation in human-specific ARHGAP11B causes basal progenitor amplification
- PMID: 27957544
- PMCID: PMC5142801
- DOI: 10.1126/sciadv.1601941
A single splice site mutation in human-specific ARHGAP11B causes basal progenitor amplification
Abstract
The gene ARHGAP11B promotes basal progenitor amplification and is implicated in neocortex expansion. It arose on the human evolutionary lineage by partial duplication of ARHGAP11A, which encodes a Rho guanosine triphosphatase-activating protein (RhoGAP). However, a lack of 55 nucleotides in ARHGAP11B mRNA leads to loss of RhoGAP activity by GAP domain truncation and addition of a human-specific carboxy-terminal amino acid sequence. We show that these 55 nucleotides are deleted by mRNA splicing due to a single C→G substitution that creates a novel splice donor site. We reconstructed an ancestral ARHGAP11B complementary DNA without this substitution. Ancestral ARHGAP11B exhibits RhoGAP activity but has no ability to increase basal progenitors during neocortex development. Hence, a single nucleotide substitution underlies the specific properties of ARHGAP11B that likely contributed to the evolutionary expansion of the human neocortex.
Keywords: ARHGAP11B; RhoGAP; basal progenitors; brain size; gene duplication; human evolution; human-specific gene; neocortex; neocortical expansion; neurogenesis.
Figures

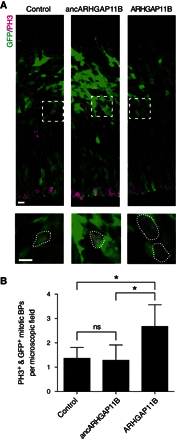
References
-
- G. F. Striedter, Principles of Brain Evolution (Sinauer Associates Inc., Sunderland, 2005).
-
- Fietz S. A., Huttner W. B., Cortical progenitor expansion, self-renewal and neurogenesis—A polarized perspective. Curr. Opin. Neurobiol. 21, 23–35 (2011). - PubMed
-
- Borrell V., Reillo I., Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 72, 955–971 (2012). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

