A genome-wide screen of bacterial mutants that enhance dauer formation in C. elegans
- PMID: 27958277
- PMCID: PMC5153853
- DOI: 10.1038/srep38764
A genome-wide screen of bacterial mutants that enhance dauer formation in C. elegans
Abstract
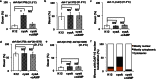
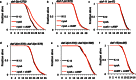
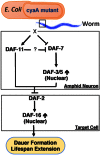
Molecular pathways involved in dauer formation, an alternate larval stage that allows Caenorhabditis elegans to survive adverse environmental conditions during development, also modulate longevity and metabolism. The decision to proceed with reproductive development or undergo diapause depends on food abundance, population density, and temperature. In recent years, the chemical identities of pheromone signals that modulate dauer entry have been characterized. However, signals derived from bacteria, the major source of nutrients for C. elegans, remain poorly characterized. To systematically identify bacterial components that influence dauer formation and aging in C. elegans, we utilized the individual gene deletion mutants in E. coli (K12). We identified 56 diverse E. coli deletion mutants that enhance dauer formation in an insulin-like receptor mutant (daf-2) background. We describe the mechanism of action of a bacterial mutant cyaA, that is defective in the production of cyclic AMP, which extends lifespan and enhances dauer formation through the modulation of TGF-β (daf-7) signaling in C. elegans. Our results demonstrate the importance of bacterial components in influencing developmental decisions and lifespan in C. elegans. Furthermore, we demonstrate that C. elegans is a useful model to study bacterial-host interactions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Golden J. W. & Riddle D. L. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218, 578–580 (1982). - PubMed
-
- Butcher R. A., Ragains J. R., Kim E. & Clardy J. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proceedings of the National Academy of Sciences of the United States of America 105, 14288–14292, doi: 10.1073/pnas.0806676105 (2008). - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

