System-specific periodicity in quantitative real-time polymerase chain reaction data questions threshold-based quantitation
- PMID: 27958340
- PMCID: PMC5154181
- DOI: 10.1038/srep38951
System-specific periodicity in quantitative real-time polymerase chain reaction data questions threshold-based quantitation
Abstract
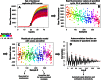
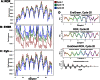
Real-time quantitative polymerase chain reaction (qPCR) data are found to display periodic patterns in the fluorescence intensity as a function of sample number for fixed cycle number. This behavior is seen for technical replicate datasets recorded on several different commercial instruments; it occurs in the baseline region and typically increases with increasing cycle number in the growth and plateau regions. Autocorrelation analysis reveals periodicities of 12 for 96-well systems and 24 for a 384-well system, indicating a correlation with block architecture. Passive dye experiments show that the effect may be from optical detector bias. Importantly, the signal periodicity manifests as periodicity in quantification cycle (Cq) values when these are estimated by the widely applied fixed threshold approach, but not when scale-insensitive markers like first- and second-derivative maxima are used. Accordingly, any scale variability in the growth curves will lead to bias in constant-threshold-based Cqs, making it mandatory that workers should either use scale-insensitive Cqs or normalize their growth curves to constant amplitude before applying the constant threshold method.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
qPCR data analysis: Better results through iconoclasm.Biomol Detect Quantif. 2019 Jun 5;17:100084. doi: 10.1016/j.bdq.2019.100084. eCollection 2019 Mar. Biomol Detect Quantif. 2019. PMID: 31194178 Free PMC article.
-
Absolute copy number from the statistics of the quantification cycle in replicate quantitative polymerase chain reaction experiments.Anal Chem. 2015 Feb 3;87(3):1889-95. doi: 10.1021/acs.analchem.5b00077. Epub 2015 Jan 23. Anal Chem. 2015. PMID: 25582662
-
Comparing real-time quantitative polymerase chain reaction analysis methods for precision, linearity, and accuracy of estimating amplification efficiency.Anal Biochem. 2014 Mar 15;449:76-82. doi: 10.1016/j.ab.2013.12.020. Epub 2013 Dec 21. Anal Biochem. 2014. PMID: 24365068
-
Bias and imprecision in analysis of real-time quantitative polymerase chain reaction data.Anal Chem. 2015 Sep 1;87(17):8925-31. doi: 10.1021/acs.analchem.5b02057. Epub 2015 Aug 13. Anal Chem. 2015. PMID: 26235706
-
[Quantitative PCR in the diagnosis of Leishmania].Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
Cited by
-
Algorithms for automated detection of hook effect-bearing amplification curves.Biomol Detect Quantif. 2018 Oct 16;16:1-4. doi: 10.1016/j.bdq.2018.08.001. eCollection 2018 Dec. Biomol Detect Quantif. 2018. PMID: 30560061 Free PMC article.
-
LoopTag FRET Probe System for Multiplex qPCR Detection of Borrelia Species.Life (Basel). 2021 Oct 31;11(11):1163. doi: 10.3390/life11111163. Life (Basel). 2021. PMID: 34833039 Free PMC article.
-
qPCR data analysis: Better results through iconoclasm.Biomol Detect Quantif. 2019 Jun 5;17:100084. doi: 10.1016/j.bdq.2019.100084. eCollection 2019 Mar. Biomol Detect Quantif. 2019. PMID: 31194178 Free PMC article.
-
Reference gene selection for quantitative real-time PCR (qRT-PCR) expression analysis in Galium aparine L.PLoS One. 2020 Feb 4;15(2):e0226668. doi: 10.1371/journal.pone.0226668. eCollection 2020. PLoS One. 2020. PMID: 32017769 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

