The ultimate and proximate mechanisms driving the evolution of long tails in forest deer mice
- PMID: 27958661
- PMCID: PMC5324611
- DOI: 10.1111/evo.13150
The ultimate and proximate mechanisms driving the evolution of long tails in forest deer mice
Abstract
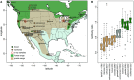
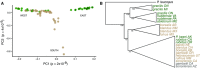
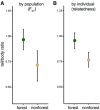
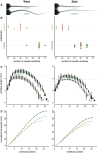
Understanding both the role of selection in driving phenotypic change and its underlying genetic basis remain major challenges in evolutionary biology. Here, we use modern tools to revisit a classic system of local adaptation in the North American deer mouse, Peromyscus maniculatus, which occupies two main habitat types: prairie and forest. Using historical collections, we find that forest-dwelling mice have longer tails than those from nonforested habitat, even when we account for individual and population relatedness. Using genome-wide SNP data, we show that mice from forested habitats in the eastern and western parts of their range form separate clades, suggesting that increased tail length evolved independently. We find that forest mice in the east and west have both more and longer caudal vertebrae, but not trunk vertebrae, than nearby prairie forms. By intercrossing prairie and forest mice, we show that the number and length of caudal vertebrae are not correlated in this recombinant population, indicating that variation in these traits is controlled by separate genetic loci. Together, these results demonstrate convergent evolution of the long-tailed forest phenotype through two distinct genetic mechanisms, affecting number and length of vertebrae, and suggest that these morphological changes-either independently or together-are adaptive.
Keywords: Caudal vertebrae; Peromyscus maniculatus; convergence; local adaptation; parallel evolution; skeletal evolution.
© 2016 The Author(s). Evolution published by Wiley Periodicals, Inc. on behalf of The Society for the Study of Evolution.
Figures

References
-
- Álvarez‐Castañeda, S. T. 2005. Peromyscus melanotis . Mammalian Species 764:1–4.
-
- Arctos: Collaborative Collections Management Solution . 2015. Available at <http://arctosdb.org>[accessed March 1, 2010].
-
- Arendt, J. , and Reznick D.. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23:26–32. - PubMed
-
- Avise, J. C. , Smith M. H., and Selander R. K.. 1979. Biochemical polymorphism and systematics in the genus Peromyscus. VII. Geographic differentiation in members of the truei and maniculatus species groups. J. Mammal. 60:177–192.
-
- Babraham Bioinformatics: Trim Galore! 0.3.7. Available at <http://www.bioinformatics.babraham.ac.uk/projects/trim_galore>
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
