Phage-inducible islands in the Gram-positive cocci
- PMID: 27959343
- PMCID: PMC5363835
- DOI: 10.1038/ismej.2016.163
Phage-inducible islands in the Gram-positive cocci
Abstract
The SaPIs are a cohesive subfamily of extremely common phage-inducible chromosomal islands (PICIs) that reside quiescently at specific att sites in the staphylococcal chromosome and are induced by helper phages to excise and replicate. They are usually packaged in small capsids composed of phage virion proteins, giving rise to very high transfer frequencies, which they enhance by interfering with helper phage reproduction. As the SaPIs represent a highly successful biological strategy, with many natural Staphylococcus aureus strains containing two or more, we assumed that similar elements would be widespread in the Gram-positive cocci. On the basis of resemblance to the paradigmatic SaPI genome, we have readily identified large cohesive families of similar elements in the lactococci and pneumococci/streptococci plus a few such elements in Enterococcus faecalis. Based on extensive ortholog analyses, we found that the PICI elements in the four different genera all represent distinct but parallel lineages, suggesting that they represent convergent evolution towards a highly successful lifestyle. We have characterized in depth the enterococcal element, EfCIV583, and have shown that it very closely resembles the SaPIs in functionality as well as in genome organization, setting the stage for expansion of the study of elements of this type. In summary, our findings greatly broaden the PICI family to include elements from at least three genera of cocci.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


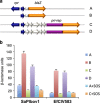
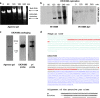
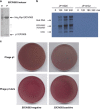

References
-
- Chen J, Novick RP. (2009). Phage-mediated intergeneric transfer of toxin genes. Science 323: 139–141. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

