Antimicrobial resistance development following surgical site infections
- PMID: 27959419
- PMCID: PMC5364857
- DOI: 10.3892/mmr.2016.6034
Antimicrobial resistance development following surgical site infections
Abstract
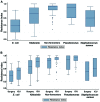
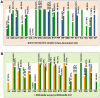
Surgical site infections (SSIs) determine an increase in hospitalization time and antibiotic therapy costs. The aim of this study was to identify the germs involved in SSIs in patients from the Clinical Emergency County Hospital of Craiova (SCJUC) and to assess their resistance to antimicrobials, with comparisons between surgical wards and the intensive care unit (ICU). The biological samples were subjected to classical bacteriological diagnostics. Antibiotic resistance was tested by disc diffusion. We used hierarchical clustering as a method to group the isolates based upon the antibiotic resistance profile. The most prevalent bacterial species isolated were Staphylococcus aureus (S. aureus; 50.72%), followed by Escherichia coli (E. coli; 17.22%) and Pseudomonas aeruginosa; 10.05%). In addition, at lower percentages, we isolated glucose-non-fermenting, Gram-negative bacteria and other Enterobacteriaceae. The antibiotic resistance varied greatly between species; the most resistant were the non-fermenting Gram‑negative rods. E. coli exhibited lower resistance to third generation cephalosporins, quinolones and carbapenems. By contrast, Klebsiella was resistant to many cephalosporins and penicillins, and to a certain extent to carbapenems due to carbapenemase production. The non-fermenting bacteria were highly resistant to antibiotics, but were generally sensitive to colistin. S. aureus was resistant to ceftriaxone (100%), penicillin (91.36%), amoxicillin/clavulanate (87.50%), amikacin (80.00%) and was sensitive to levofloxacin, doxycycline, gentamycin, tigecycline and teicoplanin. The Enterobacteriaceae resistance was only slightly higher in the ICU, particularly to carbapenems (imipenem, 31.20% in the ICU vs. 14.30% in the surgical wards; risk ratio = 2.182). As regards Staphylococcus species, but for non-fermenting bacteria, even if the median was almost the same, the antibiotic resistance index values were confined to the upper limit in the ICU. The data gathered from this study may help infection control teams to establish effective guidelines for antibiotic therapies in various surgical procedures, in order to minimize the risk of developing SSIs by the efficient application of the anti-infection armamentarium.
Figures




References
-
- Staicus C, Calina D, Rosu L, Rosu AF, Zlatian O. Involvement of microbial flora in aetiology of surgical site infections. Eur J Hosp Pharm Sci Pract. 2015;22:A59.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

