Complement Dysregulation and Disease: Insights from Contemporary Genetics
- PMID: 27959629
- PMCID: PMC6020056
- DOI: 10.1146/annurev-pathol-012615-044145
Complement Dysregulation and Disease: Insights from Contemporary Genetics
Abstract
The vertebrate complement system consists of sequentially interacting proteins that provide for a rapid and powerful host defense. Nearly 60 proteins comprise three activation pathways (classical, alternative, and lectin) and a terminal cytolytic pathway common to all. Attesting to its potency, nearly half of the system's components are engaged in its regulation. An emerging theme over the past decade is that variations in these inhibitors predispose to two scourges of modern humans. One, occurring most often in childhood, is a rare but deadly thrombomicroangiopathy called atypical hemolytic uremic syndrome. The other, age-related macular degeneration, is the most common form of blindness in the elderly. Their seemingly unrelated clinical presentations and pathologies share the common theme of overactivity of the complement system's alternative pathway. This review summarizes insights gained from contemporary genetics for understanding how dysregulation of this powerful innate immune system leads to these human diseases.
Keywords: C3; C3 glomerulopathies; CD46; age-related macular degeneration; alternative complement pathway; atypical hemolytic uremic syndrome; factor B; factor H; factor I.
Figures
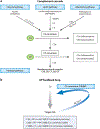


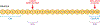

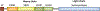
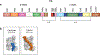

References
-
- Gros P, Milder FJ, Janssen BJ. 2008. Complement driven by conformational changes. Nat. Rev. Immunol 8:48–58 - PubMed
-
- Walport MJ. 2001. Complement. First of two parts. N. Engl. J. Med 344:1058–66 - PubMed
-
- Chaplin H, Nasongkla M, Monroe MC. 1981. Quantitation of red blood cell–bound C3d in normal subjects and random hospitalized patients. Br. J. Haematol 48(1):69–78 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

