Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors
- PMID: 27960086
- PMCID: PMC5500911
- DOI: 10.1016/j.ccell.2016.11.003
Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors
Abstract
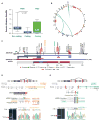
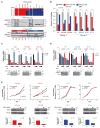
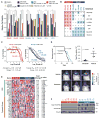
We recently reported that atypical teratoid rhabdoid tumors (ATRTs) comprise at least two transcriptional subtypes with different clinical outcomes; however, the mechanisms underlying therapeutic heterogeneity remained unclear. In this study, we analyzed 191 primary ATRTs and 10 ATRT cell lines to define the genomic and epigenomic landscape of ATRTs and identify subgroup-specific therapeutic targets. We found ATRTs segregated into three epigenetic subgroups with distinct genomic profiles, SMARCB1 genotypes, and chromatin landscape that correlated with differential cellular responses to a panel of signaling and epigenetic inhibitors. Significantly, we discovered that differential methylation of a PDGFRB-associated enhancer confers specific sensitivity of group 2 ATRT cells to dasatinib and nilotinib, and suggest that these are promising therapies for this highly lethal ATRT subtype.
Keywords: ATRT; enhancer; epigenomics; genomics; rhabdoid tumors; subgroup-specific therapeutics.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





References
-
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. - PubMed
-
- Birks DK, Donson AM, Patel PR, Dunham C, Muscat A, Algar EM, Ashley DM, Kleinschmidt-Demasters BK, Vibhakar R, Handler MH, Foreman NK. High expression of BMP pathway genes distinguishes a subset of atypical teratoid/rhabdoid tumors associated with shorter survival. Neurooncol. 2011;13:1296–1307. - PMC - PubMed
-
- Bourdeaut F, Lequin D, Brugieres L, Reynaud S, Dufour C, Doz F, Andre N, Stephan JL, Perel Y, Oberlin O, et al. Frequent hSNF5/INI1 germ-line mutations in patients with rhabdoid tumor. Clin Cancer Res. 2011;17:31–38. - PubMed
-
- Cecconi F, Proetzel G, Alvarez-Bolado G, Jay D, Gruss P. Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somitic mesoderm. Dev Dyn. 1997;210:184–190. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

