Genome analysis of canine astroviruses reveals genetic heterogeneity and suggests possible inter-species transmission
- PMID: 27965150
- PMCID: PMC7114541
- DOI: 10.1016/j.virusres.2016.12.005
Genome analysis of canine astroviruses reveals genetic heterogeneity and suggests possible inter-species transmission
Abstract
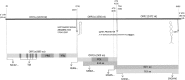
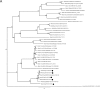
Canine astrovirus RNA was detected in the stools of 17/63 (26.9%) samples, using either a broadly reactive consensus RT-PCR for astroviruses or random RT-PCR coupled with massive deep sequencing. The complete or nearly complete genome sequence of five canine astroviruses was reconstructed that allowed mapping the genome organization and to investigate the genetic diversity of these viruses. The genome was about 6.6kb in length and contained three open reading frames (ORFs) flanked by a 5' UTR, and a 3' UTR plus a poly-A tail. ORF1a and ORF1b overlapped by 43 nucleotides while the ORF2 overlapped by 8 nucleotides with the 3' end of ORF1b. Upon genome comparison, four strains (HUN/2012/2, HUN/2012/6, HUN/2012/115, and HUN/2012/135) were more related genetically to each other and to UK canine astroviruses (88-96% nt identity), whilst strain HUN/2012/126 was more divergent (75-76% nt identity). In the ORF1b and ORF2, strains HUN/2012/2, HUN/2012/6, and HUN/2012/135 were related genetically to other canine astroviruses identified formerly in Europe and China, whereas strain HUN/2012/126 was related genetically to a divergent canine astrovirus strain, ITA/2010/Zoid. For one canine astrovirus, HUN/2012/8, only a 3.2kb portion of the genome, at the 3' end, could be determined. Interestingly, this strain possessed unique genetic signatures (including a longer ORF1b/ORF2 overlap and a longer 3'UTR) and it was divergent in both ORF1b and ORF2 from all other canine astroviruses, with the highest nucleotide sequence identity (68% and 63%, respectively) to a mink astrovirus, thus suggesting a possible event of interspecies transmission. The genetic heterogeneity of canine astroviruses may pose a challenge for the diagnostics and for future prophylaxis strategies.
Keywords: Astroviridae; Dog; Hungary; Semiconductor sequencing.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Prevalence and genome characteristics of canine astrovirus in southwest China.J Gen Virol. 2018 Jul;99(7):880-889. doi: 10.1099/jgv.0.001077. J Gen Virol. 2018. PMID: 29846155
-
Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination.Infect Genet Evol. 2017 Jun;50:38-48. doi: 10.1016/j.meegid.2017.02.008. Epub 2017 Feb 9. Infect Genet Evol. 2017. PMID: 28189887
-
Detection of a mammalian-like astrovirus in bird, European roller (Coracias garrulus).Infect Genet Evol. 2015 Aug;34:114-21. doi: 10.1016/j.meegid.2015.06.020. Epub 2015 Jun 18. Infect Genet Evol. 2015. PMID: 26096774
-
The Broad Host Range and Genetic Diversity of Mammalian and Avian Astroviruses.Viruses. 2017 May 10;9(5):102. doi: 10.3390/v9050102. Viruses. 2017. PMID: 28489047 Free PMC article. Review.
-
Novel human astroviruses: Novel human diseases?J Clin Virol. 2016 Sep;82:56-63. doi: 10.1016/j.jcv.2016.07.004. Epub 2016 Jul 11. J Clin Virol. 2016. PMID: 27434149 Review.
Cited by
-
Phylogenetic Analysis of Lednice Orthobunyavirus.Microorganisms. 2019 Oct 13;7(10):447. doi: 10.3390/microorganisms7100447. Microorganisms. 2019. PMID: 31614950 Free PMC article.
-
Detection and characterisation of canine astrovirus, canine parvovirus and canine papillomavirus in puppies using next generation sequencing.Sci Rep. 2019 Mar 14;9(1):4602. doi: 10.1038/s41598-019-41045-z. Sci Rep. 2019. PMID: 30872719 Free PMC article.
-
Molecular survey of parvovirus, astrovirus, coronavirus, and calicivirus in symptomatic dogs.Vet Res Commun. 2021 Feb;45(1):31-40. doi: 10.1007/s11259-020-09785-w. Epub 2021 Jan 4. Vet Res Commun. 2021. PMID: 33392909 Free PMC article.
-
Metatranscriptomic identification of novel RNA viruses from raccoon dog (Nyctereutes procyonoides) feces in Japan.Sci Rep. 2025 Feb 27;15(1):7100. doi: 10.1038/s41598-025-90474-6. Sci Rep. 2025. PMID: 40016305 Free PMC article.
-
Detection and phylogenetic characterization of astroviruses in insectivorous bats from Central-Southern Italy.Zoonoses Public Health. 2018 Sep;65(6):702-710. doi: 10.1111/zph.12484. Epub 2018 Jun 12. Zoonoses Public Health. 2018. PMID: 29896884 Free PMC article.
References
-
- Bosch A., Guix S., Krishna N., Mendez E., Monroe S.S., Pantin-Jackwood M., Schultz-Cherry S. 2010. Nineteen New Species in the Genus Mamastrovirus in the Astroviridae Family. (ICTV 2010018aV2010b)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

