Rapid Production of Virus Protein Microarray Using Protein Microarray Fabrication through Gene Synthesis (PAGES)
- PMID: 27965274
- PMCID: PMC5294215
- DOI: 10.1074/mcp.M116.064873
Rapid Production of Virus Protein Microarray Using Protein Microarray Fabrication through Gene Synthesis (PAGES)
Abstract
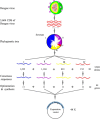
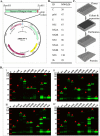
The high genetic variability of RNA viruses is a significant factor limiting the discovery of effective biomarkers, the development of vaccines, and characterizations of the immune response during infection. Protein microarrays have been shown to be a powerful method in biomarker discovery and the identification of novel protein-protein interaction networks, suggesting that this technique could also be very useful in studies of infectious RNA viruses. However, to date, the amount of genetic material required to produce protein arrays, as well as the time- and labor-intensive procedures typically needed, have limited their more widespread application. Here, we introduce a method, protein microarray fabrication through gene synthesis (PAGES), for the rapid and efficient construction of protein microarrays particularly for RNA viruses. Using dengue virus as an example, we first identify consensus sequences from 3,604 different strains and then fabricate complete proteomic microarrays that are unique for each consensus sequence. To demonstrate their applicability, we show that these microarrays can differentiate sera from patients infected by dengue virus, related pathogens, or from uninfected patients. We anticipate that the microarray and expression library constructed in this study will find immediate use in further studies of dengue virus and that, more generally, PAGES will become a widely applied method in the clinical characterization of RNA viruses.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Complete nucleotide sequence analysis of a Dengue-1 virus isolated on Easter Island, Chile.Arch Virol. 2008;153(10):1967-70. doi: 10.1007/s00705-008-0200-0. Epub 2008 Sep 25. Arch Virol. 2008. PMID: 18815724
-
Increasing Clinical Severity during a Dengue Virus Type 3 Cuban Epidemic: Deep Sequencing of Evolving Viral Populations.J Virol. 2016 Apr 14;90(9):4320-4333. doi: 10.1128/JVI.02647-15. Print 2016 May. J Virol. 2016. PMID: 26889031 Free PMC article.
-
An "on-matrix" digestion procedure for AP-MS experiments dissects the interplay between complex-conserved and serotype-specific reactivities in Dengue virus-human plasma interactome.J Proteomics. 2019 Feb 20;193:71-84. doi: 10.1016/j.jprot.2017.07.004. Epub 2017 Jul 13. J Proteomics. 2019. PMID: 28713027
-
Advances in the development of human protein microarrays.Expert Rev Proteomics. 2017 Jul;14(7):627-641. doi: 10.1080/14789450.2017.1347042. Expert Rev Proteomics. 2017. PMID: 28644690 Review.
-
Establishment and Application of Flavivirus Replicons.Adv Exp Med Biol. 2018;1062:165-173. doi: 10.1007/978-981-10-8727-1_12. Adv Exp Med Biol. 2018. PMID: 29845532 Review.
Cited by
-
Multiplexed Biomarker Panels Discriminate Zika and Dengue Virus Infection in Humans.Mol Cell Proteomics. 2018 Feb;17(2):349-356. doi: 10.1074/mcp.RA117.000310. Epub 2017 Nov 15. Mol Cell Proteomics. 2018. PMID: 29141913 Free PMC article.
-
Advances in Virus Biorecognition and Detection Techniques for the Surveillance and Prevention of Infectious Diseases.Biosensors (Basel). 2025 Mar 20;15(3):198. doi: 10.3390/bios15030198. Biosensors (Basel). 2025. PMID: 40136995 Free PMC article. Review.
-
The N and C-terminal deleted variant of the dengue virus NS1 protein is a potential candidate for dengue vaccine development.Sci Rep. 2024 Aug 14;14(1):18883. doi: 10.1038/s41598-024-65593-1. Sci Rep. 2024. PMID: 39143088 Free PMC article.
-
Glycosaminoglycan microarrays for studying glycosaminoglycan-protein systems.Carbohydr Polym. 2024 Jul 1;335:122106. doi: 10.1016/j.carbpol.2024.122106. Epub 2024 Mar 29. Carbohydr Polym. 2024. PMID: 38616080 Free PMC article. Review.
-
Developments and Applications of Functional Protein Microarrays.Mol Cell Proteomics. 2020 Jun;19(6):916-927. doi: 10.1074/mcp.R120.001936. Epub 2020 Apr 17. Mol Cell Proteomics. 2020. PMID: 32303587 Free PMC article. Review.
References
-
- Gire S. K., Goba A., Andersen K. G., Sealfon R. S., Park D. J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G., Wohl S., Moses L. M., Yozwiak N. L., Winnicki S., Matranga C. B., Malboeuf C. M., Qu J., Gladden A. D., Schaffner S. F., Yang X., Jiang P. P., Nekoui M., Colubri A., Coomber M. R., Fonnie M., Moigboi A., Gbakie M., Kamara F. K., Tucker V., Konuwa E., Saffa S., Sellu J., Jalloh A. A., Kovoma A., Koninga J., Mustapha I., Kargbo K., Foday M., Yillah M., Kanneh F., Robert W., Massally J. L., Chapman S. B., Bochicchio J., Murphy C., Nusbaum C., Young S., Birren B. W., Grant D. S., Scheiffelin J. S., Lander E. S., Happi C., Gevao S. M., Gnirke A., Rambaut A., Garry R. F., Khan S. H., and Sabeti P. C. (2014) Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 - PMC - PubMed
-
- Cohen J. (2016) The race for a Zika vaccine is on. Science 351, 543–544 - PubMed
-
- Normile D. (2014) Tropical diseases. Dengue vaccine trial poses public health quandary. Science 345, 367–368 - PubMed
-
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., and Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

