Structure of the Lipid Nanodisc-reconstituted Vacuolar ATPase Proton Channel: DEFINITION OF THE INTERACTION OF ROTOR AND STATOR AND IMPLICATIONS FOR ENZYME REGULATION BY REVERSIBLE DISSOCIATION
- PMID: 27965356
- PMCID: PMC5290949
- DOI: 10.1074/jbc.M116.766790
Structure of the Lipid Nanodisc-reconstituted Vacuolar ATPase Proton Channel: DEFINITION OF THE INTERACTION OF ROTOR AND STATOR AND IMPLICATIONS FOR ENZYME REGULATION BY REVERSIBLE DISSOCIATION
Abstract
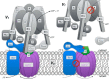
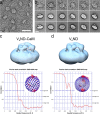
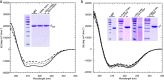
Eukaryotic vacuolar H+-ATPase (V-ATPase) is a multisubunit enzyme complex that acidifies subcellular organelles and the extracellular space. V-ATPase consists of soluble V1-ATPase and membrane-integral Vo proton channel sectors. To investigate the mechanism of V-ATPase regulation by reversible disassembly, we recently determined a cryo-EM reconstruction of yeast Vo The structure indicated that, when V1 is released from Vo, the N-terminal cytoplasmic domain of subunit a (aNT) changes conformation to bind rotor subunit d However, insufficient resolution precluded a precise definition of the aNT-d interface. Here we reconstituted Vo into lipid nanodiscs for single-particle EM. 3D reconstructions calculated at ∼15-Å resolution revealed two sites of contact between aNT and d that are mediated by highly conserved charged residues. Alanine mutagenesis of some of these residues disrupted the aNT-d interaction, as shown by isothermal titration calorimetry and gel filtration of recombinant subunits. A recent cryo-EM study of holo V-ATPase revealed three major conformations corresponding to three rotational states of the central rotor of the enzyme. Comparison of the three V-ATPase conformations with the structure of nanodisc-bound Vo revealed that Vo is halted in rotational state 3. Combined with our prior work that showed autoinhibited V1-ATPase to be arrested in state 2, we propose a model in which the conformational mismatch between free V1 and Vo functions to prevent unintended reassembly of holo V-ATPase when activity is not needed.
Keywords: EM; Vo proton channel; isothermal titration calorimetry (ITC); lipid nanodisc; protein structure; proton transport; vacuolar ATPase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

