Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression
- PMID: 27965449
- PMCID: PMC5156302
- DOI: 10.1128/mBio.01790-16
Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression
Abstract
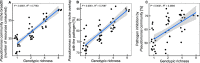
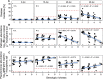
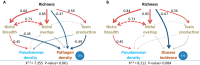
Bacterial communities associated with plant roots play an important role in the suppression of soil-borne pathogens, and multispecies probiotic consortia may enhance disease suppression efficacy. Here we introduced defined Pseudomonas species consortia into naturally complex microbial communities and measured the importance of Pseudomonas community diversity for their survival and the suppression of the bacterial plant pathogen Ralstonia solanacearum in the tomato rhizosphere microbiome. The survival of introduced Pseudomonas consortia increased with increasing diversity. Further, high Pseudomonas diversity reduced pathogen density in the rhizosphere and decreased the disease incidence due to both intensified resource competition and interference with the pathogen. These results provide novel mechanistic insights into elevated pathogen suppression by diverse probiotic consortia in naturally diverse plant rhizospheres. Ecologically based community assembly rules could thus play a key role in engineering functionally reliable microbiome applications.
Importance: The increasing demand for food supply requires more-efficient control of plant diseases. The use of probiotics, i.e., naturally occurring bacterial antagonists and competitors that suppress pathogens, has recently reemerged as a promising alternative to agrochemical use. It is, however, still unclear how many and which strains we should choose for constructing effective probiotic consortia. Here we present a general ecological framework for assembling effective probiotic communities based on in vitro characterization of community functioning. Specifically, we show that increasing the diversity of probiotic consortia enhances community survival in the naturally diverse rhizosphere microbiome, leading to increased pathogen suppression via intensified resource competition and interference with the pathogen. We propose that these ecological guidelines can be put to the test in microbiome engineering more widely in the future.
Copyright © 2016 Hu et al.
Figures
Similar articles
-
Phages enhance both phytopathogen density control and rhizosphere microbiome suppressiveness.mBio. 2024 Jun 12;15(6):e0301623. doi: 10.1128/mbio.03016-23. Epub 2024 May 23. mBio. 2024. PMID: 38780276 Free PMC article.
-
Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health.Nat Commun. 2015 Sep 24;6:8413. doi: 10.1038/ncomms9413. Nat Commun. 2015. PMID: 26400552 Free PMC article.
-
Introduction of probiotic bacterial consortia promotes plant growth via impacts on the resident rhizosphere microbiome.Proc Biol Sci. 2021 Oct 13;288(1960):20211396. doi: 10.1098/rspb.2021.1396. Epub 2021 Oct 13. Proc Biol Sci. 2021. PMID: 34641724 Free PMC article.
-
The rhizosphere microbiome and plant health.Trends Plant Sci. 2012 Aug;17(8):478-86. doi: 10.1016/j.tplants.2012.04.001. Epub 2012 May 5. Trends Plant Sci. 2012. PMID: 22564542 Review.
-
Microbial diversity and interactions: Synergistic effects and potential applications of Pseudomonas and Bacillus consortia.Microbiol Res. 2025 Apr;293:128054. doi: 10.1016/j.micres.2025.128054. Epub 2025 Jan 7. Microbiol Res. 2025. PMID: 39799763 Review.
Cited by
-
Natural Cross-Kingdom Spread of Apple Scar Skin Viroid from Apple Trees to Fungi.Cells. 2022 Nov 20;11(22):3686. doi: 10.3390/cells11223686. Cells. 2022. PMID: 36429116 Free PMC article.
-
Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety.Foods. 2021 Mar 12;10(3):602. doi: 10.3390/foods10030602. Foods. 2021. PMID: 33809159 Free PMC article. Review.
-
Combining Different Potato-Associated Pseudomonas Strains for Improved Biocontrol of Phytophthora infestans.Front Microbiol. 2018 Oct 29;9:2573. doi: 10.3389/fmicb.2018.02573. eCollection 2018. Front Microbiol. 2018. PMID: 30420845 Free PMC article.
-
The biocontrol potentials of rhizospheric bacterium Bacillus velezensis K0T24 against mulberry bacterial wilt disease.Arch Microbiol. 2024 Apr 15;206(5):213. doi: 10.1007/s00203-024-03935-3. Arch Microbiol. 2024. PMID: 38616201
-
Microbe to Microbiome: A Paradigm Shift in the Application of Microorganisms for Sustainable Agriculture.Front Microbiol. 2020 Dec 21;11:622926. doi: 10.3389/fmicb.2020.622926. eCollection 2020. Front Microbiol. 2020. PMID: 33408712 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
