Ameloblastoma RNA profiling uncovers a distinct non-coding RNA signature
- PMID: 27965463
- PMCID: PMC5354851
- DOI: 10.18632/oncotarget.13889
Ameloblastoma RNA profiling uncovers a distinct non-coding RNA signature
Abstract
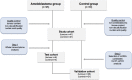
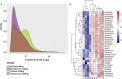
Ameloblastoma of the jaws remains the top difficult to treat odontogenic tumour and has a high recurrence rate. New evidence suggests that non-coding RNAs (ncRNAs) play a critical role in tumourgenesis and prognosis of cancer. However, ameloblastoma ncRNA expression data is lacking. Here we present the first report of ameloblastoma ncRNA signatures. A total of 95 ameloblastoma cases and a global array transcriptome technology covering > 285.000 full-length transcripts were used in this two-step analysis. The analysis first identified in a test cohort 31 upregulated ameloblastoma-associated ncRNAs accompanied by signalling pathways of cancer, spliceosome, mRNA surveillance and Wnt. Further validation in an independent cohort points out the long non-coding (lncRNAs) and small nucleolar RNA (snoRNAs): LINC340, SNORD116-25, SNORA11, SNORA21, SNORA47 and SNORA65 as a distinct ncRNA signature of ameloblastoma. Importantly, the presence of these ncRNAs was independent of BRAF-V600E and SMO-L412F mutations, histology type or tumour location, but was positively correlated with the tumour size. Taken together, this study shows a systematic investigation of ncRNA expression of ameloblastoma, and illuminates new diagnostic and therapeutic targets for this invasive odontogenic tumour.
Keywords: ameloblastoma; biomarkers; gene expression analysis; ncRNA; transcriptome.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

