Genome Sequencing of Sulfolobus sp. A20 from Costa Rica and Comparative Analyses of the Putative Pathways of Carbon, Nitrogen, and Sulfur Metabolism in Various Sulfolobus Strains
- PMID: 27965637
- PMCID: PMC5127849
- DOI: 10.3389/fmicb.2016.01902
Genome Sequencing of Sulfolobus sp. A20 from Costa Rica and Comparative Analyses of the Putative Pathways of Carbon, Nitrogen, and Sulfur Metabolism in Various Sulfolobus Strains
Abstract
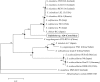
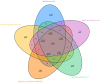
The genome of Sulfolobus sp. A20 isolated from a hot spring in Costa Rica was sequenced. This circular genome of the strain is 2,688,317 bp in size and 34.8% in G+C content, and contains 2591 open reading frames (ORFs). Strain A20 shares ~95.6% identity at the 16S rRNA gene sequence level and <30% DNA-DNA hybridization (DDH) values with the most closely related known Sulfolobus species (i.e., Sulfolobus islandicus and Sulfolobus solfataricus), suggesting that it represents a novel Sulfolobus species. Comparison of the genome of strain A20 with those of the type strains of S. solfataricus, Sulfolobus acidocaldarius, S. islandicus, and Sulfolobus tokodaii, which were isolated from geographically separated areas, identified 1801 genes conserved among all Sulfolobus species analyzed (core genes). Comparative genome analyses show that central carbon metabolism in Sulfolobus is highly conserved, and enzymes involved in the Entner-Doudoroff pathway, the tricarboxylic acid cycle and the CO2 fixation pathways are predominantly encoded by the core genes. All Sulfolobus species encode genes required for the conversion of ammonium into glutamate/glutamine. Some Sulfolobus strains have gained the ability to utilize additional nitrogen source such as nitrate (i.e., S. islandicus strain REY15A, LAL14/1, M14.25, and M16.27) or urea (i.e., S. islandicus HEV10/4, S. tokodaii strain7, and S. metallicus DSM 6482). The strategies for sulfur metabolism are most diverse and least understood. S. tokodaii encodes sulfur oxygenase/reductase (SOR), whereas both S. islandicus and S. solfataricus contain genes for sulfur reductase (SRE). However, neither SOR nor SRE genes exist in the genome of strain A20, raising the possibility that an unknown pathway for the utilization of elemental sulfur may be present in the strain. The ability of Sulfolobus to utilize nitrate or sulfur is encoded by a gene cluster flanked by IS elements or their remnants. These clusters appear to have become fixed at a specific genomic site in some strains and lost in other strains during the course of evolution. The versatility in nitrogen and sulfur metabolism may represent adaptation of Sulfolobus to thriving in different habitats.
Keywords: Sulfolobus; carbon metabolism; comparative genomics; genome sequencing; nitrogen metabolism; strain A20; sulfur metabolism.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

