Molecular Cloning, Characterization, and Expression of MiSOC1: A Homolog of the Flowering Gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from Mango (Mangifera indica L)
- PMID: 27965680
- PMCID: PMC5126060
- DOI: 10.3389/fpls.2016.01758
Molecular Cloning, Characterization, and Expression of MiSOC1: A Homolog of the Flowering Gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from Mango (Mangifera indica L)
Abstract
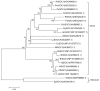
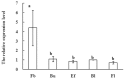
MADS-box transcription factor plays a crucial role in plant development, especially controlling the formation and development of floral organs. Mango (Mangifera indica L) is an economically important fruit crop, but its molecular control of flowering is largely unknown. To better understand the molecular basis of flowering regulation in mango, we isolated and characterized the MiSOC1, a putative mango orthologs for the Arabidopsis SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1/AGAMOUS-LIKE 20 (SOC1/AGL20) with homology-based cloning and RACE. The full-length cDNA (GenBank accession No.: KP404094) is 945 bp in length including a 74 bp long 5' UTR and a 189 bp long 3' UTR and the open reading frame was 733 bps, encoding 223 amino acids with molecular weight 25.6 kD. Both sequence alignment and phylogenetic analysis all indicated that deduced protein contained a conservative MADS-box and semi-conservative K domain and belonged to the SOC1/TM3 subfamily of the MADS-box family. Quantitative real-time PCR was performed to investigate the expression profiles of MiSOC1 gene in different tissues/organs including root, stem, leaves, flower bud, and flower. The result indicated MiSOC1 was widely expressed at different levels in both vegetative and reproductive tissues/organs with the highest expression level in the stems' leaves and inflorescences, low expression in roots and flowers. The expression of MiSOC1 in different flower developmental stages was different while same tissue -specific pattern among different varieties. In addition, MiSOC1 gene expression was affect by ethephon while high concentration ethephon inhibit the expression of MiSOC1. Overexpression of MiSOC1 resulted in early flowering in Arabidopsis. In conclusion, these results suggest that MiSOC1 may act as induce flower function in mango.
Keywords: MADS-box; MiSOC1; analysis; flowering; mango (Mangifera indica L).
Figures






Similar articles
-
Genome-wide analysis of the MADS-box gene family in mango and ectopic expression of MiMADS77 in Arabidopsis results in early flowering.Gene. 2025 Jan 30;935:149054. doi: 10.1016/j.gene.2024.149054. Epub 2024 Oct 28. Gene. 2025. PMID: 39490648
-
Identification and characterization of FaSOC1, a homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from strawberry.Gene. 2013 Dec 1;531(2):158-67. doi: 10.1016/j.gene.2013.09.036. Epub 2013 Sep 17. Gene. 2013. PMID: 24055423
-
Ectopic expression of two CAULIFLOWER genes from mango caused early flowering in Arabidopsis.Gene. 2023 Jan 30;851:146931. doi: 10.1016/j.gene.2022.146931. Epub 2022 Oct 13. Gene. 2023. PMID: 36244548
-
Overexpression of two DELLA subfamily genes MiSLR1 and MiSLR2 from mango promotes early flowering and enhances abiotic stress tolerance in Arabidopsis.Plant Sci. 2024 Dec;349:112242. doi: 10.1016/j.plantsci.2024.112242. Epub 2024 Sep 5. Plant Sci. 2024. PMID: 39244094
-
The roles of non-structural carbohydrates in fruiting: a review focusing on mango (Mangifera indica).Funct Plant Biol. 2024 Apr;51:FP23195. doi: 10.1071/FP23195. Funct Plant Biol. 2024. PMID: 38588720 Review.
Cited by
-
GhSOC1s Evolve to Respond Differently to the Environmental Cues and Promote Flowering in Partially Independent Ways.Front Plant Sci. 2022 Apr 20;13:882946. doi: 10.3389/fpls.2022.882946. eCollection 2022. Front Plant Sci. 2022. PMID: 35519808 Free PMC article.
-
Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango.Int J Mol Sci. 2021 Sep 10;22(18):9802. doi: 10.3390/ijms22189802. Int J Mol Sci. 2021. PMID: 34575962 Free PMC article.
-
Isolation and Functional Characterization of Two CONSTANS-like 16 (MiCOL16) Genes from Mango.Int J Mol Sci. 2022 Mar 12;23(6):3075. doi: 10.3390/ijms23063075. Int J Mol Sci. 2022. PMID: 35328495 Free PMC article.
-
Isolation and Functional Characterization of a LEAFY Gene in Mango (Mangifera indica L.).Int J Mol Sci. 2022 Apr 2;23(7):3974. doi: 10.3390/ijms23073974. Int J Mol Sci. 2022. PMID: 35409334 Free PMC article.
-
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 influences flowering time, lateral branching, oil quality, and seed yield in Brassica juncea cv. Varuna.Funct Integr Genomics. 2019 Jan;19(1):43-60. doi: 10.1007/s10142-018-0626-8. Epub 2018 Jun 25. Funct Integr Genomics. 2019. PMID: 29943206
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

